22 July 2021: Database Analysis
Identification of Key Genes and Underlying Mechanisms in Acute Kawasaki Disease Based on Bioinformatics Analysis
Side Gao1BCDEF, Wenjian Ma1BCD, Xuze Lin1BCD, Sizhuang Huang1BC, Mengyue Yu1AG*DOI: 10.12659/MSM.930547
Med Sci Monit 2021; 27:e930547
Abstract
BACKGROUND: Kawasaki disease (KD) is a systemic vasculitis that predominantly occurs in children, but the pathogenesis of KD remains unclear. Here, we explored key genes and underlying mechanisms potentially involved in KD using bioinformatic analyses.
MATERIAL AND METHODS: The shared differentially expressed genes (DEGs) in KD compared to control samples were identified using the microarray data from the Gene Expression Omnibus Series (GSE) 18606, GSE68004, and GSE73461. Analyses of the functional annotation, protein-protein interaction (PPI) network, microRNA-target DEGs regulatory network, and immune cell infiltration were performed. The expression of hub genes before and after intravenous immunoglobulin (IVIG) treatment in KD was further verified using GSE16797.
RESULTS: A total of 195 shared DEGs (164 upregulated and 31 downregulated genes) were identified between KD and healthy controls. These shared DEGs were mainly enriched in immune and inflammatory responses. Ten upregulated hub genes (ITGAX, SPI1, LILRB2, MMP9, S100A12, C3AR1, RETN, MAPK14, TLR5, MYD88) and the most significant module were identified in the PPI network. There were 309 regulatory relationships detected within 70 predicted microRNAs and 193 target DEGs. The immune cell infiltration analysis showed that monocytes, neutrophils, activated mast cells, and activated natural killer cells had relatively high proportions and were significantly more infiltrated in KD samples. Six hub genes of ITGAX, LILRB2, C3AR1, MAPK14, TLR5, and MYD88 were markedly downregulated after IVIG treatment for KD.
CONCLUSIONS: Our study identified the candidate genes and associated molecules that may be related to the KD process, and provided new insights into potential mechanisms and therapeutic targets for KD.
Keywords: Databases, Genetic, Diagnosis, Mucocutaneous Lymph Node Syndrome, Computational Biology, Immunoglobulins, Microarray Analysis, Protein Interaction Maps
Background
Kawasaki disease (KD), also called mucocutaneous lymph node syndrome, is an acute, self-limited febrile illness of unknown causes that predominantly affects children below 5 years of age [1]. It is a multisystem disorder characterized by acute vasculitis in small and medium-sized arteries, especially in coronary arteries [2]. This disease is more markedly prevalent in children in Japan (annual incidence 264.8/100 000 in 2012), whereas the incidence was estimated to be 19/100 000 in the USA and 8.4/100 000 in the UK in children below 5 years of age [3,4]. As reported, the incidence of KD is 2.5 times higher in Asians and Pacific Islanders and 1.5 times higher in Blacks compared to Whites [4]. The course of KD needs more attention since nearly 25% of people with KD, if left untreated, will experience vascular complications such as coronary artery aneurysm (CAA), thromboembolism, stenosis, or even sudden death [5]. KD has been reported as the leading cause of acquired heart diseases in children in developed countries [5]. Although timely treatment with intravenous immunoglobulin (IVIG) reduces the risk of CAA, some children who do not respond to IVIG are still at considerable risks of developing coronary artery damage [6]. In addition, the lack of specific diagnostic tests and biomarkers for KD make early diagnosis and treatment challenging. Therefore, KD remains an important focus of research and it is extremely important to perform detailed exploration of the etiopathogenesis of KD and to further improve healthcare for this population.
The etiology of KD has been studied extensively, but the exact pathogenic factors or triggering agents for KD are still unidentified, and the underlying mechanisms for initiation and progression of KD remain largely unknown. Recently, it has been suggested that KD involves the complex interplay of infectious triggers and genetic susceptibility followed by an abnormal immune response [5]. A large number of candidate pathogens has been tested and discarded. Some case reports linked KD with viral agents; however, no specific infectious agent has been shown to be consistently and causally associated with KD [6]. Genetic factors also increase susceptibility to KD, as suggested by differences in incidence of KD among races [6]. Moreover, KD occurs as a consequence of a dysregulated immune system, which involves the activation and infiltration of the arterial wall by cells of both the innate and adaptive immune systems [7]. Recent studies have found that several genes are closely associated with the KD process [8–10], indicating that genetic alterations may play a critical role in the development of KD.
In this study, we identified the shared differentially expressed genes (DEGs) and underlying pathways in acute KD compared to healthy control samples among 3 GSE datasets by using an integrated bioinformatic analysis. Base on the shared DEGs, analyses of functional annotation, protein-protein interaction (PPI) network, microRNA (miRNA)-DEGs network construction, and immune cell infiltration were performed. We also verified the hub gene expression in KD before and after IVIG treatment. With these analyses, we hope to identify biomarkers for early KD detection and provide potential therapeutic targets for KD.
Material and Methods
MICROARRAY DATA RESOURCES:
The microarray data of GSE18606, GSE68004, and GSE73461 comparing gene expression in whole-blood samples between acute KD and healthy controls were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) to screen out DEGs involved in KD. Another GSE16797 dataset was also obtained from the GEO database and used to validate the hub gene expression before and after IVIG treatment in acute KD patients. Detailed information on these datasets is shown in Table 1. Study flowchart is shown in Figure 1A.
IDENTIFICATION OF DEGS:
Raw gene expression data were read and processed using affy package (version 1.50.0) in R software (version 4.0.1) [11]. Gene probes were annotated using an annotation profile provided by the platform, and unmatched probes were discarded. When multiple probes matched one gene symbol, the average values of probes were calculated as the final expression of the gene. DEGs between KD and control samples in each GSE dataset were screened out using Linear Models for Microarray (Limma) package (Version 3.30.3) in R, and the threshold was defined as |log2(foldchange)| >1 and P<0.05. DEGs were labeled as upregulated and downregulated. The significance of DEGs in each dataset was exhibited in a volcano plot using ggplot2 in R. To visualize the shared DEGs among the 3 datasets, the online tool Bioinformatics & Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to draw Venn diagrams. Further, the differences in shared DEGs are shown in heatmaps created using pheatmap in R.
FUNCTIONAL ANNOTATION:
Biological functions of the shared DEGs were explored with the Gene Ontology (GO) [12] and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses [13] using the online tool Database for Annotation, Visualization, and integrated discovery (DAVID) (Version 6.8; https://david.ncifcrf.gov/home.jsp) [14]. The GO terms included biological process (BP), cellular component (CC), and molecular function (MF). Significant GO and KEGG pathways with the thresholds of count ≥2 and P<0.05 were selected for further analysis.
PROTEIN–PROTEIN INTERACTION (PPI) NETWORK:
The PPI network among shared DEGs was constructed using the Search tool for the retrieval of interacting genes/proteins (STRING) database (Version 11.0; www.string-db.org) [15], and a PPI score (medium confidence) ≥0.4 was defined as the cutoff value. The network was then visualized using Cytoscape (Version 3.7.1; www.cytoscape.org) [16]. The degree of each node, considered as the significance of DEGs-encoded protein in the network, was calculated using the cytoHubba plugin in Cytoscape [17]. DEGs with the top 10 highest degrees were selected as hub genes. Moreover, the Molecular Complex Detection (MCODE) plugin in Cytoscape was used to identify significant modules in the network [18], and the parameters were set as follows: Degree Cutoff=2, Node Score Cutoff=0.2, K-Core=2, Max. Depth=100. The most common and largest module was defined as MCODE score >5.
CONSTRUCTION OF MIRNA-TARGET DEGS REGULATORY NETWORKS:
The potential miRNAs of shared DEGs were predicted based on miRTarBase using Enrichr (http://amp.pharm.mssm.edu/Enrichr/) [19] with the threshold of P<0.05. A regulatory network between predicted miRNA and target DEGs was constructed using Cytoscape.
IMMUNE CELL INFILTRATION ANALYSIS:
CIBERSORT is a recently developed algorithm used for sensitive and specific quantification of the relative levels of 22 human immune cell phenotypes within a complex gene expression mixture (https://cibersort.stanford.edu) [20]. Normalized expression profiles of shared DEGs in all the KD and control samples among the 3 datasets were extracted and used to calculate the immune infiltration scores. The samples were screened according to P<0.05, and the percentage of each type of immune cells in samples (immune infiltration score) was calculated using the CIBERSORT algorithm. The deviations of immune infiltration in each sample are shown in a stacked bar chart, while the differences in immune infiltration levels of each immune cell type between control and KD groups are shown in a violin graph generated via R software.
VALIDATION OF HUB GENE EXPRESSION IN KD BEFORE AND AFTER IVIG TREATMENT:
To investigate if key genes would be differently expressed before and after IVIG treatment in KD, we validated the hub gene expression among pre- and post-IVIG samples using another GSE16797 dataset. Box charts were plotted using GraphPad prism (Version 8.0.2).
Results
IDENTIFICATION OF DEGS IN KD:
A total of 195 shared DEGs were identified in the whole blood of acute KD pediatric patients (before treatment) compared to control samples (Figure 1B), including 164 upregulated and 31 downregulated genes. The foldchange and significance of DEGs in each dataset are shown in Figure 2. The heatmap of the shared DEGs indicated that these genes could easily distinguish KD from controls (Figure 3), suggesting that they may play a critical role in the development of KD.
A detailed list of shared DEGs is shown in Supplementary Table 1.
FUNCTIONAL ENRICHMENT ANALYSES OF SHARED DEGS:
The GO and KEGG enrichment analyses revealed that the BP of shared DEGs were mainly enriched in immune and inflammatory responses, and cell surface receptor signaling pathway; the CC were mainly existed in extracellular exosome and plasma membrane; and the MF were mostly enriched in receptor activity (Figure 4A–4C). Meanwhile, the significantly enriched KEGG pathways of shared DEGs included osteoclast differentiation, hematopoietic cell lineage, tumor necrosis factor signaling pathway, and immune-related diseases (Figure 4D). The top 10 results of GO-BP and KEGG enrichment analyses are shown in Supplementary Tables 2 and 3.
PPI NETWORK ANALYSIS:
A total of 157 nodes and 555 interactional pairs were included in the PPI network (Figure 5A). After calculating degrees via cytoHubba, DEGs with the top 10 highest degrees were selected as hub genes, including ITGAX, SPI1, LILRB2, MMP9, S100A12, C3AR1, RETN, MAPK14, TLR5, and MYD88. They were all significantly upregulated in KD and their functions are summarized in Table 2. Further, the most significant module in the PPI network with 23 nodes and 126 edges was identified (MCODE score=11.4), and the hub genes of ITGAX, RETN, and LILRB2 were also included (Figure 5B).
MIRNA-TARGET DEGS REGULATORY NETWORK ANALYSIS:
In the miRNA-target DEGs network, there were 309 regulatory relationships detected within 263 nodes, including 70 predicted miRNAs and 193 DEGs (Figure 6). The top 3 most significant miRNAs were hsa-miR-34a-5p (P=1.08E-05), has-miR-122-3p (P=7.82E-04) and has-miR-548ah-5p (P=0.001), respectively.
IMMUNE CELL INFILTRATION ANALYSIS:
The differences in immune infiltration between the KD and control samples were analyzed by the CIBERSORT algorithm. As a result, the 22 subtypes of immune cells were both detected in KD and controls, and the immunocyte composition of the entire cohort is summarized via a box plot in Figure 7A. The violin plot indicated that 17 types of immune cells were differentially infiltrated between KD and controls (Figure 7B). Specifically, plasma cells, γδ T cells, activated natural killer (NK) cells, monocytes, resting dendritic cells (DCs), resting and activated mast cells, and neutrophils had higher infiltration in KD samples, while naïve B cells, naïve CD4 T cells, resting and activated memory CD4 T cells, resting NK cells, M1 and M2 macrophages, activated DCs, and eosinophils had relatively lower infiltration in KD than that in the control group. Of note, the monocytes, neutrophils, activated mast cells, and activated NK cells had relatively high proportions and were significantly more infiltrated in KD samples, suggesting that they may play a central role in the pathogenesis of acute KD.
VALIDATION OF HUB GENE EXPRESSION IN KD BEFORE AND AFTER IVIG TREATMENT:
We focused on the effect of IVIG of hub gene expression in KD patients. The results showed that, out of the 10 hub genes, 6 genes were differentially expressed among pre- and post-IVIG KD samples, including ITGAX, LILRB2, C3AR1, MAPK14, TLR5, and MYD88. These genes were originally upregulated in acute KD as compared with control, while after IVIG infusion, they were markedly downregulated and back to normal levels (Figure 8). This indicated that these 6 hub genes were closely related to immune and inflammatory responses in KD and might be potential therapeutic targets for IVIG treatment.
Discussion
It is reported that not only infectious stimulus and immune responses are responsible for the occurrence of KD, but that genetic susceptibility also plays an essential role [2]. In Japan, there is a 10-fold higher risk of KD in siblings of patients with KD than in the general population and a 2-fold higher risk in those with parental history of KD [21]. These differences in the family and races suggest that genetic factors might be involved in the pathogenesis of KD. Previous studies have found that single-nucleotide polymorphisms (SNPs) in multiple genes are associated with KD and disease outcome (such as aneurysm formation and response to IVIG), including 1,4,5-triphosphate 3-kinase (ITPKC), caspase-3 (CASP3), B lymphocyte kinase (BLK), CD40, and human leukocyte antigen (HLA) [8–10]. Genetic variants in transforming growth factor (TGF)-β pathways also contributes to increased risk of aneurysm formation in European patients [22]. In addition to polymorphisms and mutations, altered gene expression is also a promoter in KD [23]; however, mechanisms linking the genetic alterations with KD progression are still poorly elucidated and require more investigation.
With the aid of the large open-sourced GEO database, bioinformatic analyses based on gene microarray profiles have been used to reveal the key hub genes and potential therapeutic targets of multiple diseases. However, related investigations in KD based on gene and molecular levels are still lacking. Herein, we obtained the genome-wide expression data in whole-blood samples of patients with acute KD and in controls using 3 different GSE datasets, and performed an integrated analysis. As a result, 195 shared DEGs were screened out and they were mainly enriched in immune and inflammatory responses. The identified 10 hub genes were all significantly upregulated in KD. Some of them have been found to be closely related to KD, while the function of the other genes in KD warrants further research. We also detected potential miRNAs involved in KD, analyzed the immune cell infiltration in KD samples, and validated the hub gene expression after IVIG treatment. All these data may help us better understand the etiology of KD.
The 10 hub genes (
Increased expression and activity of a diverse set of MMPs have been demonstrated in acute KD [24]. The expression of MMP3 and MMP9, both known to mediate vascular smooth muscle cell (VSMC) migration and neointimal formation, are increased in patients with KD, and the circulating levels of these MMPs are correlated with the formation of CAAs in KD [25–27]. MMP9 also was reported to lead to elastin breakdown in a mouse model of KD [28]. Among the S100 protein family, serum concentrations of S100A12 are substantially higher in acute KD and decline after IVIG treatment [29–31], while in patients with giant CAAs, plasma S100A12 levels remain elevated, highlighting its potential utility as a biomarker to monitor long-term persistence of arteritis [30]. S100A12 also promotes vasculitis by stimulating monocytes to produce interleukin (IL)-1β, which directly induces coronary artery endothelial cell dysfunction [31]. Post-mortem studies have revealed that 73% of KD patients have acute kidney injury involving glomerulonephritis with deposition of IgA and complement C3 [32]. The deposition of C3 can enhance the damage of small and medium-sized vessels, particularly the coronary arteries [33]. C3a, along with its receptor, C3aR, can also induce a variety of vascular inflammation and remodeling [34]. As for resistin, it is reported that the circulating resistin levels are significantly higher in patients with KD and are correlated with the occurrence of CAAs [35]. Resistin also enhances inflammation by cross-talking with the nuclear factor-κB in a mouse coronary arteritis model [36]. The TLR5 is a key member of TLRs, which may stimulate the immunopathogenesis of KD and vasculitis [37]. Interestingly, we verified the hub gene expression in KD before and after IVIG treatment, and found that the hub genes of
Immune responses are closely related to the pathogenesis of KD, involving activation and infiltration of both innate and adaptive immune cells. These cells can release proinflammatory cytokines such as IL-1β and TNF-α, and further enhance inflammation and promote necrotizing arteritis [38]. Their matric products also progressively obstruct the coronary lumen and induce a variety of cardiovascular lesions [38]. Consistently, we found that the monocytes, neutrophils, activated mast cells, and activated NK cells mainly compose the inflammatory infiltrate in KD samples and they were significantly upregulated compared with controls. These data indicate that these immune cell infiltrations play an important role in the progression of KD vasculitis.
miRNA is a class of small non-coding RNAs that regulate mRNA expression. Of human coding genes, 60–70% are estimated to be regulated by miRNAs, which are emerging as critical gene regulators in a host of cellular processes, including apoptosis, inflammation, and innate immune response [39]. Additionally, circulating miRNAs are easily detectable, relatively stable, and tissue-specific, making them attractive disease biomarkers [40]. Several studies have found that the miRNA profiles of serum exosome or coronary artery tissues are associated with acute KD, including miR-23a, miR-27b, miR-223, and miR-145 [41–42]. In the present study, we also identified 70 predicted miRNAs that may be potentially related to KD, of which hsa-miR-34a-5p had the most target DEGs and the most significant value. It is reported that miR-34a-5p can upregulate the IL-1β-mediated inflammatory process, while inhibiting miR-34a-5p attenuates the lipopolysaccharide-induced injury of endothelial cells [43,44]. We assumed that miR-34a-5p might serve as a diagnostic biomarker for KD. However, these predicted miRNAs have not been validated in a KD cohort and their functional roles require further experiments.
Several limitations should be acknowledged in this study. First, the shared DEGs in KD were identified via microarray data re-analysis. Further in vitro and in vivo validation experiments of cell lines and animal models are needed to reveal their specific functions in the pathogenesis of KD. Second, the incidence of KD varies based on ethnicity, but the KD samples of the GSE datasets were limited to patients from the USA, UK, and Japan, leading to a potential racial bias. Thus, our results may not be universally applicable and should be verified by international, multi-center, and large-scale studies. Third, the prognostic power of the identified DEGs of KD remains unclear. Therefore, the performance of these genes for risk stratification and outcomes prediction in KD patients warrants further investigations.
Conclusions
The present study identified the key genes and underlying mechanisms involved in Kawasaki disease, which may provide new perspectives to reveal the molecular pathogenesis of Kawasaki disease and develop potential diagnostic and therapeutic targets.
Figures
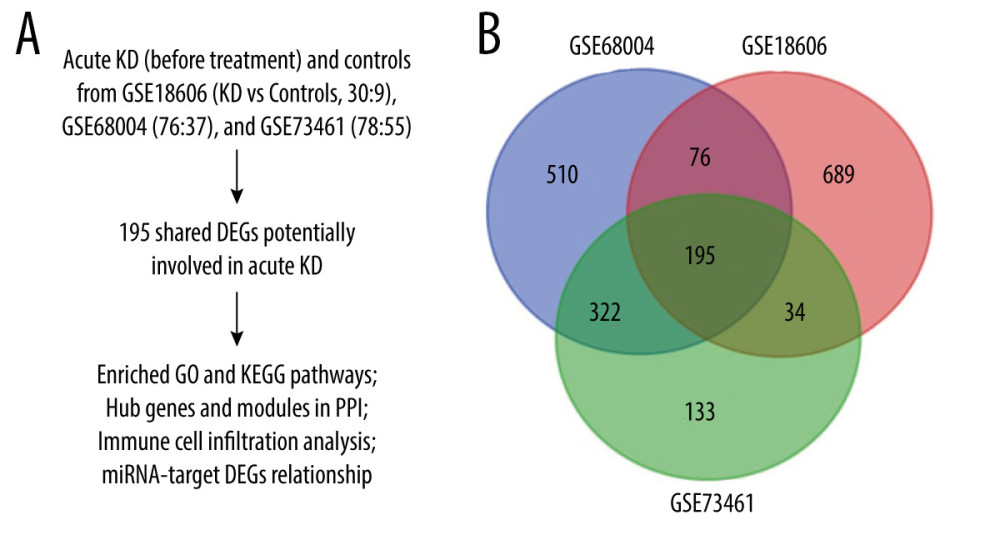 Figure 1. Study flowchart and Venn diagram. (A) The GSE datasets and bioinformatic analyses used in this study. (B) Venn diagram showing the 195 shared DEGs in GSE18606, GSE68004, and GSE73461.
Figure 1. Study flowchart and Venn diagram. (A) The GSE datasets and bioinformatic analyses used in this study. (B) Venn diagram showing the 195 shared DEGs in GSE18606, GSE68004, and GSE73461. 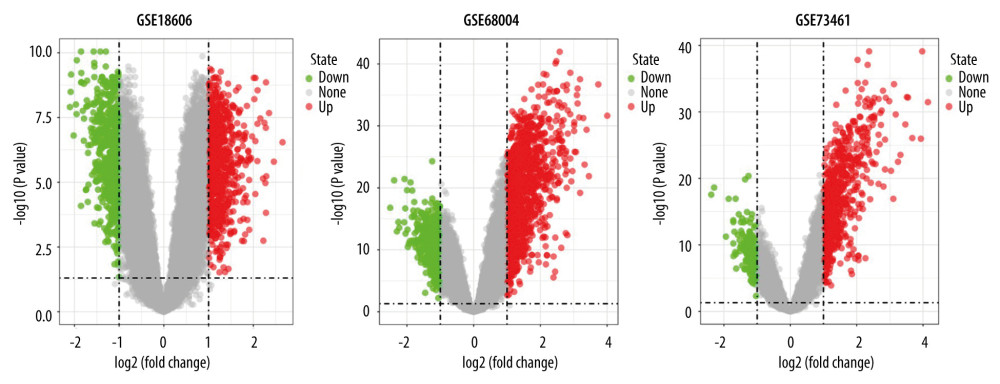 Figure 2. Volcano plots showing the DEGs in each GSE dataset. The x-axis was log2 (foldchange) and y-axis was −log10 (P value) in volcano plots. The red dots indicate upregulated genes, and green dots indicate downregulated genes.
Figure 2. Volcano plots showing the DEGs in each GSE dataset. The x-axis was log2 (foldchange) and y-axis was −log10 (P value) in volcano plots. The red dots indicate upregulated genes, and green dots indicate downregulated genes. 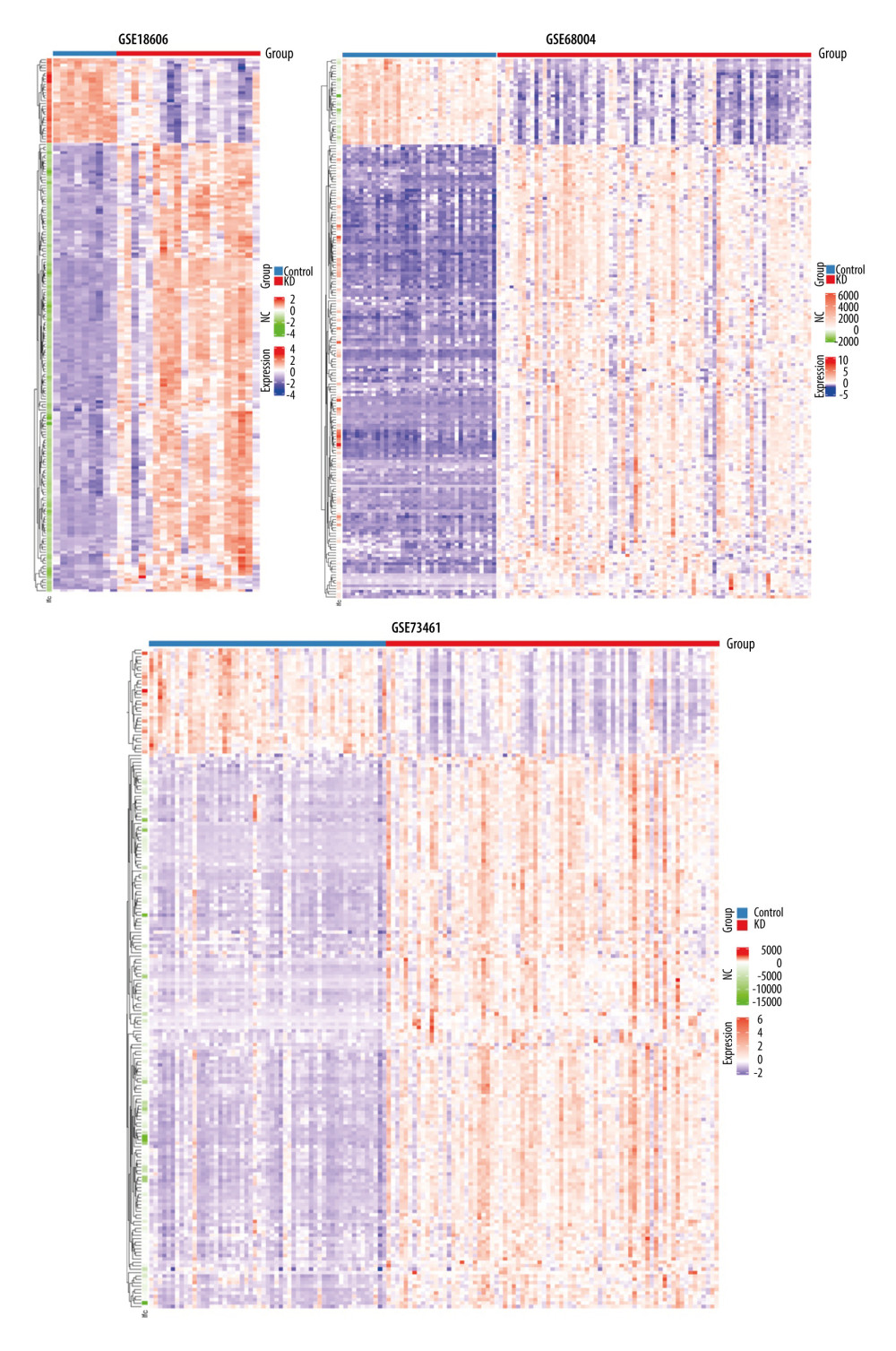 Figure 3. Heatmaps of the shared DEGs in the 3 datasets. The expression of the 195 shared DEGs among KD and controls in each dataset are shown via heatmap. The x-axis represents different samples, and y-axis represents different genes. The red boxes indicate upregulated genes, and blue boxes indicate downregulated genes.
Figure 3. Heatmaps of the shared DEGs in the 3 datasets. The expression of the 195 shared DEGs among KD and controls in each dataset are shown via heatmap. The x-axis represents different samples, and y-axis represents different genes. The red boxes indicate upregulated genes, and blue boxes indicate downregulated genes. 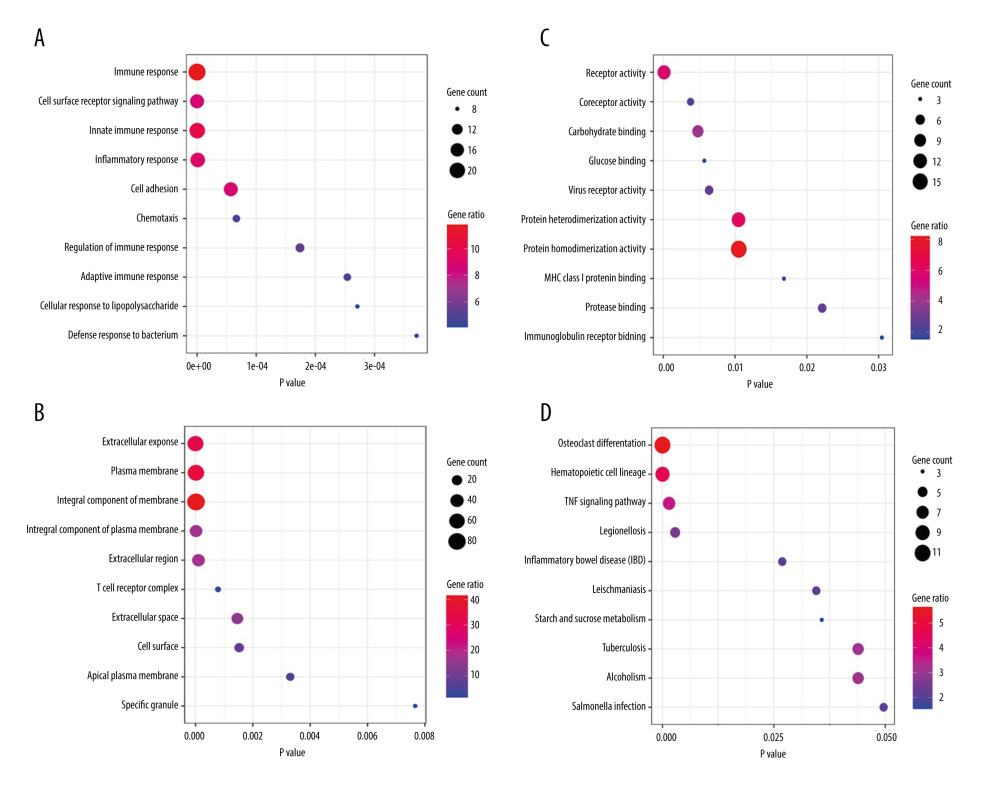 Figure 4. GO and KEGG enrichment analyses of shared DEGs. GO enrichment analyses of top 10 most significantly enriched biological process (A), cellular component (B), and molecular function (C) of the shared DEGs, and the top 10 enriched KEGG pathways of the shared DEGs (D).
Figure 4. GO and KEGG enrichment analyses of shared DEGs. GO enrichment analyses of top 10 most significantly enriched biological process (A), cellular component (B), and molecular function (C) of the shared DEGs, and the top 10 enriched KEGG pathways of the shared DEGs (D). 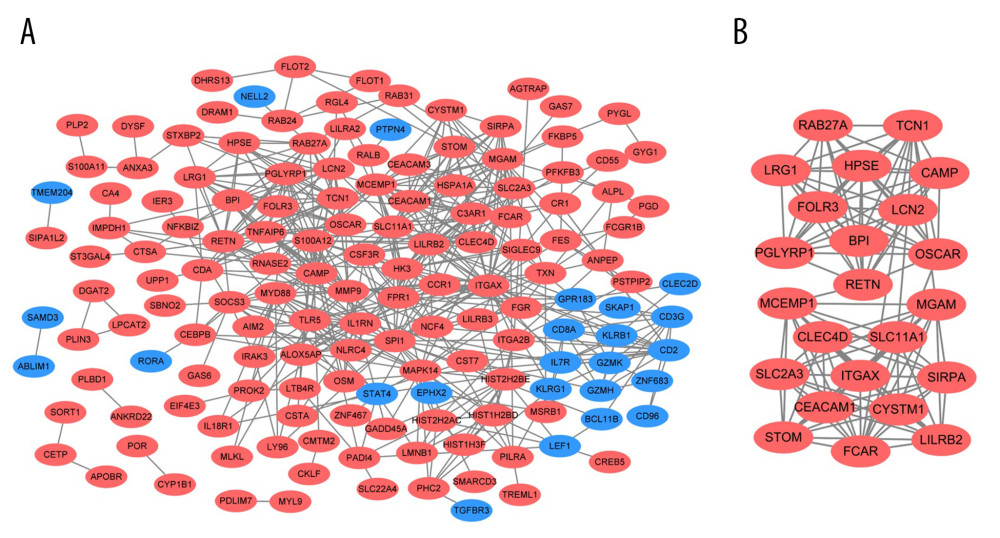 Figure 5. PPI network and the most significant module. Upregulated genes are marked in red and downregulated genes are marked in blue in the PPI network (A). The most significant module in PPI network was also identified (B).
Figure 5. PPI network and the most significant module. Upregulated genes are marked in red and downregulated genes are marked in blue in the PPI network (A). The most significant module in PPI network was also identified (B). 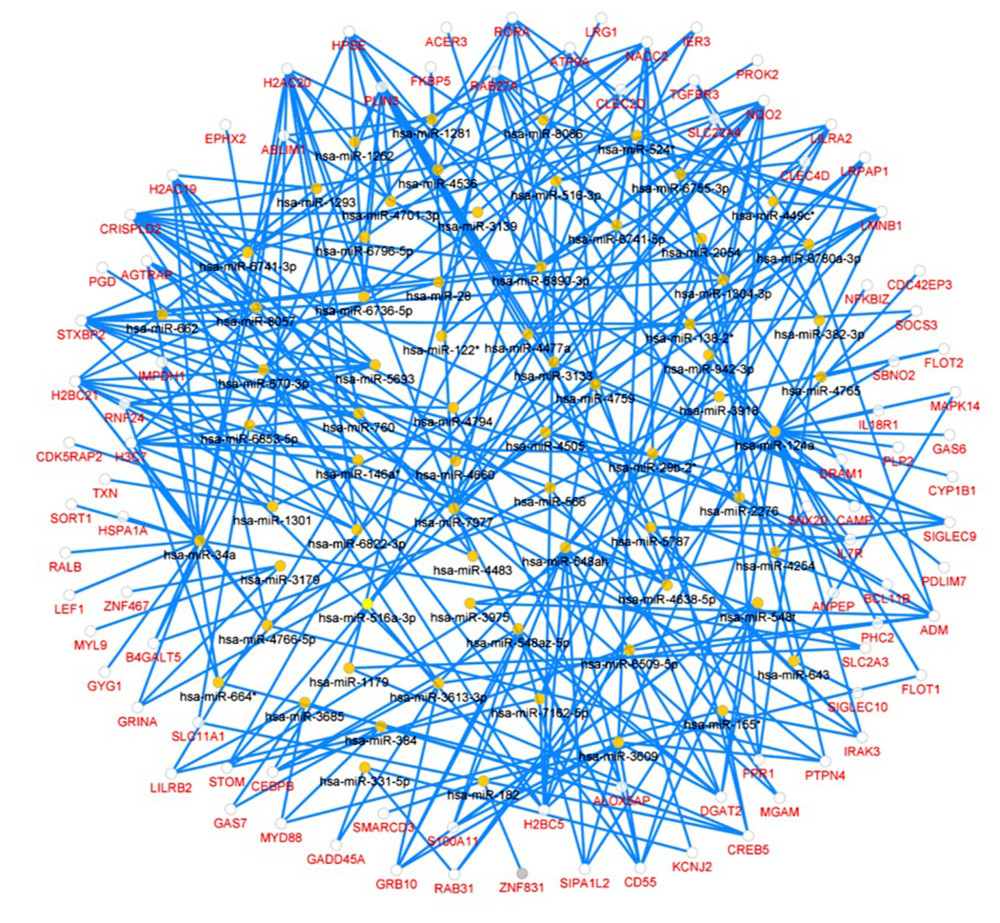 Figure 6. Predicted miRNA-target DEGs regulatory network. Regulatory network between predicted miRNA and target DEGs. The red dots indicate DEGs and yellow dots indicate miRNAs. The blue lines indicate miRNA-DEGs pairs.
Figure 6. Predicted miRNA-target DEGs regulatory network. Regulatory network between predicted miRNA and target DEGs. The red dots indicate DEGs and yellow dots indicate miRNAs. The blue lines indicate miRNA-DEGs pairs. 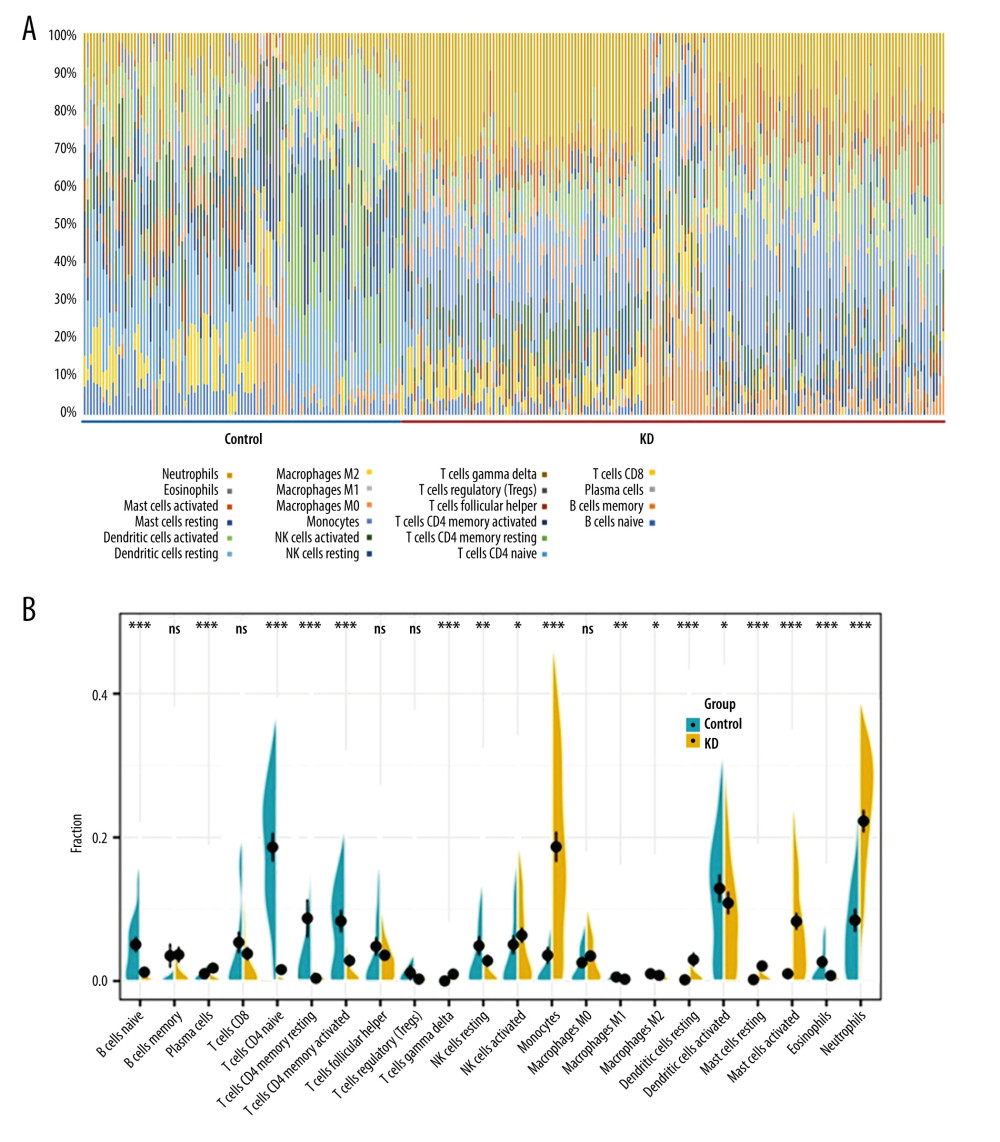 Figure 7. Immune cell infiltration analysis of the shared DEGs showing the infiltration levels of 22 immune cell subtypes in KD and controls. (A) Stacked bar chart showing deviations of immune infiltration in each control or KD sample among the 3 datasets. (B) Violin plot showing differences in proportions of each immune cell type between control and KD groups. * P<0.05, ** P<0.01, *** P<0.001, ns: not significant.
Figure 7. Immune cell infiltration analysis of the shared DEGs showing the infiltration levels of 22 immune cell subtypes in KD and controls. (A) Stacked bar chart showing deviations of immune infiltration in each control or KD sample among the 3 datasets. (B) Violin plot showing differences in proportions of each immune cell type between control and KD groups. * P<0.05, ** P<0.01, *** P<0.001, ns: not significant. 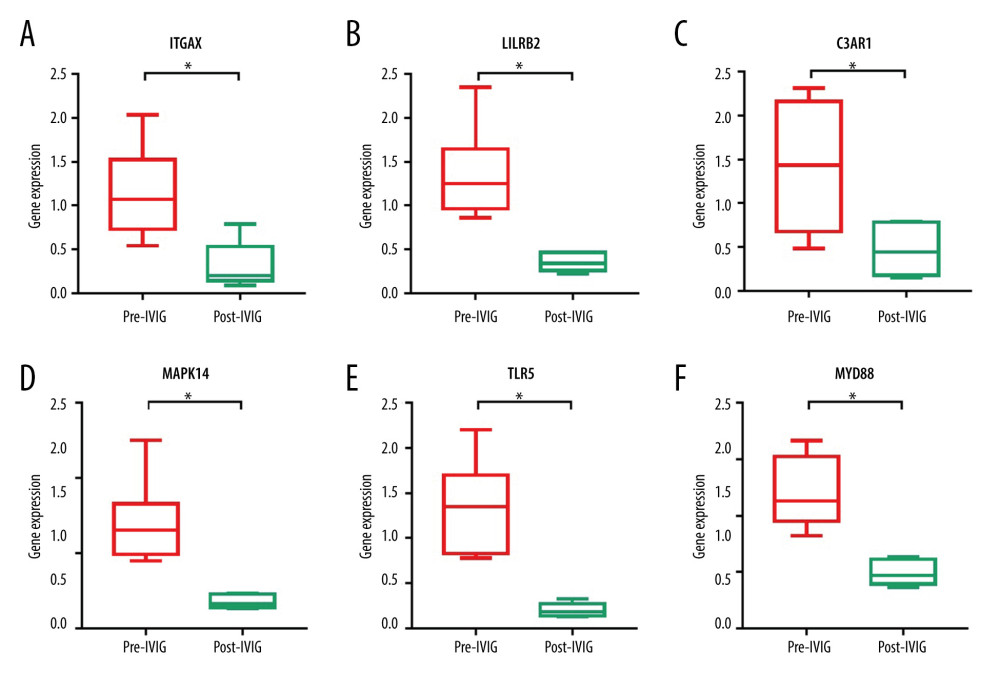 Figure 8. Validation of the hub gene expression changes in KD before and after IVIG treatment. Expression of the identified 10 hub genes in pre- and post-IVIG treatment KD were compared using GSE16797, and 6 genes were significantly downregulated after IVIG. (A) ITGAX. (B) LILRB2. (C) C3AR1. (D) MAPK14. (E) TLR5. (F) MYD88. The x-axis shows different groups and y-axis shows a log2 transformation of gene expression. * P<0.05, ** P<0.01.
Figure 8. Validation of the hub gene expression changes in KD before and after IVIG treatment. Expression of the identified 10 hub genes in pre- and post-IVIG treatment KD were compared using GSE16797, and 6 genes were significantly downregulated after IVIG. (A) ITGAX. (B) LILRB2. (C) C3AR1. (D) MAPK14. (E) TLR5. (F) MYD88. The x-axis shows different groups and y-axis shows a log2 transformation of gene expression. * P<0.05, ** P<0.01. Tables
Table 1. Gene expression datasets used in this study. Table 2. The hub genes in Kawasaki disease.
Table 2. The hub genes in Kawasaki disease.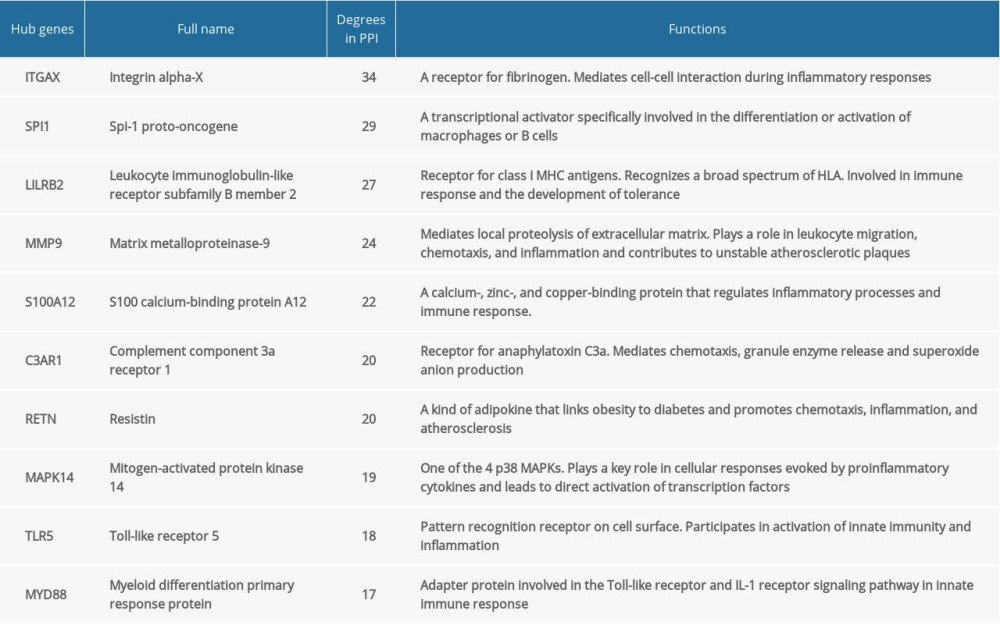 Supplementary Table 1. Screening of 195 shared DEGs in KD.
Supplementary Table 1. Screening of 195 shared DEGs in KD. Supplementary Table 2. The top 10 results of GO-BP enrichment analyses of the shared DEGs.
Supplementary Table 2. The top 10 results of GO-BP enrichment analyses of the shared DEGs.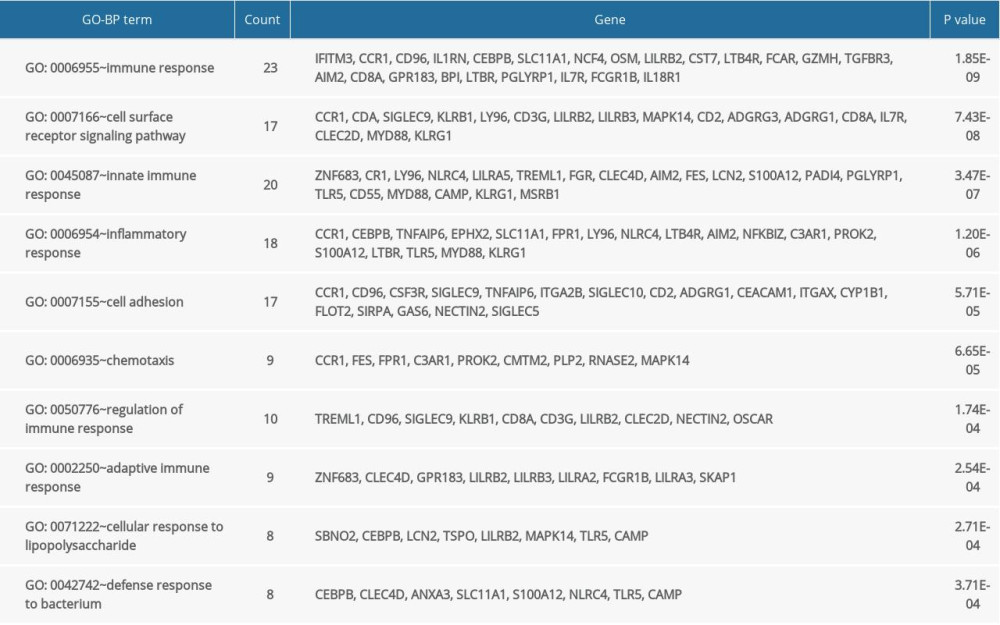 Supplementary Table 3. The top 10 results of KEGG pathway analyses of the shared DEGs.
Supplementary Table 3. The top 10 results of KEGG pathway analyses of the shared DEGs.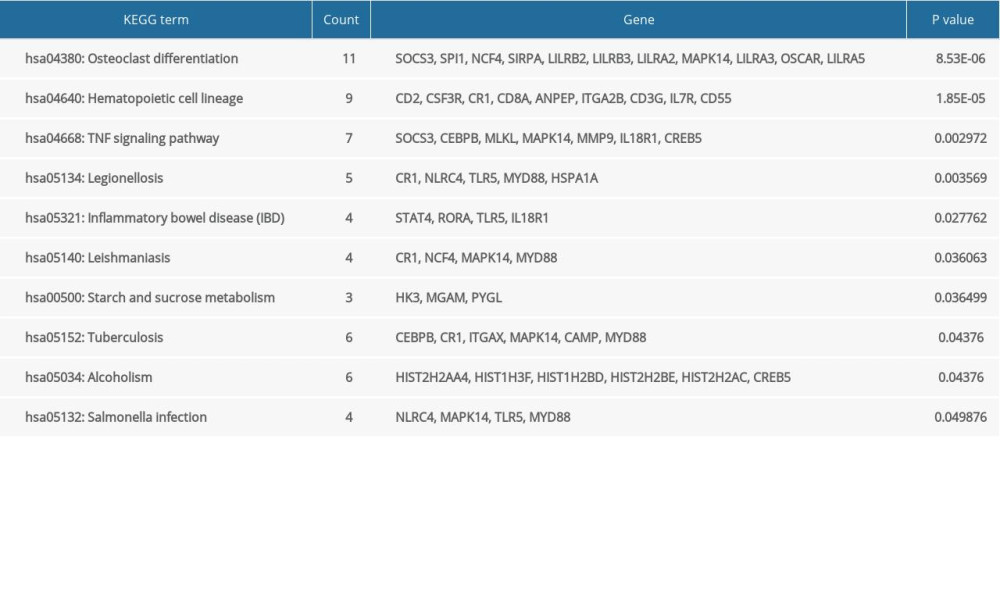
References
1. Dietz SM, van Stijn D, Burgner D, Dissecting Kawasaki disease: A state-of-the-art review: Eur J Pediatr, 2017; 176(8); 995-1009
2. Dimitriades VR, Brown AG, Gedalia A, Kawasaki disease: Pathophysiology, clinical manifestations, and management: Curr Rheumatol Rep, 2014; 16(6); 423
3. Holman RC, Christensen KY, Belay ED, Racial/ethnic differences in the incidence of Kawasaki syndrome among children in Hawaii: Hawaii Med J, 2010; 69; 194-97
4. Uehara R, Belay ED, Epidemiology of Kawasaki disease in Asia, Europe, and the United States: J Epidemiol, 2012; 22(2); 79-85
5. McCrindle BW, Rowley AH, Newburger JW, Diagnosis, treatment, and long-term management of Kawasaki disease: A scientific statement for health professionals from the American Heart Association: Circulation, 2017; 135(17); e927-99
6. Agarwal S, Agrawal DK, Kawasaki disease: Etiopathogenesis and novel treatment strategies: Expert Rev Clin Immunol, 2017; 13(3); 247-58
7. Noval Rivas M, Arditi M, Kawasaki disease: Pathophysiology and insights from mouse models: Nat Rev Rheumatol, 2020; 16(7); 391-405
8. Onouchi Y, Gunji T, Burns JC, ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms: Nat Genet, 2008; 40; 35-42
9. Onouchi Y, Ozaki K, Buns JC, Common variants in CASP3 confer susceptibility to Kawasaki disease: Hum Mol Genet, 2010; 19; 2898-906
10. Onouchi Y, Ozaki K, Burns JC, A genome-wide association study identifies three new risk loci for Kawasaki disease: Nat Genet, 2012; 44; 517-21
11. Irizarry RA, Hobbs B, Collin F, Exploration, normalization, and summaries of high-density oligonucleotide array probe level data: Biostatistics, 2003; 4(2); 249-64
12. Ashburner M, Ball CA, Blake JA, Gene Ontology: Tool for the unification of biology: Nat Genet, 2000; 25(1); 25-29
13. Kanehisa M, Goto S, KEGG: Kyoto encyclopedia of genes and genomes: Nucleic Acids Res, 2000; 28(1); 27-30
14. Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources: Nat Protoc, 2009; 4(1); 44-57
15. Szklarczyk D, Franceschini A, Wyder S, STRING v10: Protein–protein interaction networks, integrated over the tree of life: Nucleic Acids Res, 2015; 43; D447-52
16. Shannon P, Markiel A, Ozier O, Cytoscape: A software environment for integrated models of biomolecular interaction networks: Genome Res, 2003; 13(11); 2498-504
17. Chin CH, Chen SH, Wu HH, cytoHubba: Identifying hub objects and sub-networks from complex interactome: BMC Syst Biol, 2014; 8(Suppl 4); S11
18. Bader GD, Hogue CW, An automated method for finding molecular complexes in large protein interaction networks: BMC Bioinformatics, 2003; 4; 2
19. Chen EY, Tan CM, Kou Y, Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool: BMC Bioinformatics, 2013; 14; 128
20. Newman AM, Liu CL, Green MR, Robust enumeration of cell subsets from tissue expression profiles: Nat Methods, 2015; 12(5); 453-57
21. Takahashi K, Oharaseki T, Yokouchi Y, Update on etiology and immunopathogenesis of Kawasaki disease: Curr Opin Rheumatol, 2014; 26(1); 31-36
22. Shimizu C, Jain S, Davila S, Transforming growth factor-beta signaling pathway in patients with Kawasaki disease: Circ Cardiovasc Genet, 2011; 4; 16-25
23. Popper SJ, Shimizu C, Shike H, Gene-expression patterns reveal underlying biological processes in Kawasaki disease: Genome Biol, 2007; 8(12); R261
24. Senzaki H, The pathophysiology of coronary artery aneurysms in Kawasaki disease: Role of matrix metalloproteinases: Arch Dis Child, 2006; 91(10); 847-51
25. Shimizu C, Matsubara T, Onouchi Y, Matrix metalloproteinase haplotypes associated with coronary artery aneurysm formation in patients with Kawasaki disease: J Hum Genet, 2010; 55(12); 779-84
26. Korematsu S, Ohta Y, Tamai N, Cell distribution differences of matrix metalloproteinase-9 and tissue inhibitor of matrix metalloproteinase-1 in patients with Kawasaki disease: Pediatr Infect Dis J, 2012; 31(9); 973-74
27. Johnson JL, Dwivedi A, Somerville M, Matrix metalloproteinase (MMP)-3 activates MMP-9 mediated vascular smooth muscle cell migration and neointima formation in mice: Arterioscler Thromb Vasc Biol, 2011; 31(9); e35-44
28. Lau AC, Duong TT, Ito S, Matrix metalloproteinase 9 activity leads to elastin breakdown in an animal model of Kawasaki disease: Arthritis Rheum, 2008; 58(3); 854-63
29. Abe J, Jibiki T, Noma S, Gene expression profiling of the effect of high-dose intravenous Ig in patients with Kawasaki disease: J Immunol, 2005; 174(9); 5837-45
30. Lech M, Guess J, Duffner J, Circulating markers of inflammation persist in children and adults with giant aneurysms after Kawasaki disease: Circ Genom Precis Med, 2019; 12(4); e002433
31. Armaroli G, Verweyen E, Pretzer C, Monocyte-derived interleukin-1β as the driver of S100A12-induced sterile inflammatory activation of human coronary artery endothelial cells: Implications for the pathogenesis of Kawasaki disease: Arthritis Rheumatol, 2019; 71(5); 792-804
32. Takahashi K, Oharaseki T, Yokouchi Y, Kawasaki disease as a systemic vasculitis in childhood: Ann Vasc Dis, 2010; 3(3); 173-81
33. Watanabe T, Clinical features of acute kidney injury in patients with Kawasaki disease: World J Clin Pediatr, 2018; 7(3); 83-88
34. Hovland A, Jonasson L, Garred P, The complement system and toll-like receptors as integrated players in the pathophysiology of atherosclerosis: Atherosclerosis, 2015; 241(2); 480-94
35. Cai X, Zhu Q, Wu T, Association of circulating resistin and adiponectin levels with Kawasaki disease: A meta-analysis: Exp Ther Med, 2020; 19(2); 1033-41
36. Gao F, Si F, Feng S, Resistin enhances inflammatory cytokine production in coronary artery tissues by activating the NF-κB signaling: Biomed Res Int, 2016; 2016; 3296437
37. Huang YH, Li SC, Huang LH, Identifying genetic hypomethylation and upregulation of Toll-like receptors in Kawasaki disease: Oncotarget, 2017; 8(7); 11249-58
38. Orenstein JM, Shulman ST, Fox LM, Three linked vasculopathic processes characterize Kawasaki disease: A light and transmission electron microscopic study: PLoS One, 2012; 7(6); e38998
39. Tahamtan A, Teymoori-Rad M, Nakstad B, Anti-inflammatory MicroRNAs and their potential for inflammatory diseases treatment: Front Immunol, 2018; 9; 1377
40. van Rooij E, The art of microRNA research: Circ Res, 2011; 108(2); 219-34
41. Rowley AH, Pink AJ, Reindel R, A study of cardiovascular miRNA biomarkers for Kawasaki disease: Pediatr Infect Dis J, 2014; 33(12); 1296-99
42. Kuo HC, Hsieh KS, Ming-Huey Guo M, Next-generation sequencing identifies micro-RNA-based biomarker panel for Kawasaki disease: J Allergy Clin Immunol, 2016; 138(4); 1227-30
43. Luo X, Hu R, Zheng Y, Metformin shows anti-inflammatory effects in murine macrophages through Dicer/microribonucleic acid-34a-5p and microribonucleic acid-125b-5p: J Diabetes Investig, 2020; 11(1); 101-9
44. Zhang FM, Zheng WH, Wang HJ, MiR-34a-5p inhibition attenuates LPS-induced endothelial cell injury by targeting FOXM1: Eur Rev Med Pharmacol Sci, 2020; 24(20); 10829-38
Figures
 Figure 1. Study flowchart and Venn diagram. (A) The GSE datasets and bioinformatic analyses used in this study. (B) Venn diagram showing the 195 shared DEGs in GSE18606, GSE68004, and GSE73461.
Figure 1. Study flowchart and Venn diagram. (A) The GSE datasets and bioinformatic analyses used in this study. (B) Venn diagram showing the 195 shared DEGs in GSE18606, GSE68004, and GSE73461. Figure 2. Volcano plots showing the DEGs in each GSE dataset. The x-axis was log2 (foldchange) and y-axis was −log10 (P value) in volcano plots. The red dots indicate upregulated genes, and green dots indicate downregulated genes.
Figure 2. Volcano plots showing the DEGs in each GSE dataset. The x-axis was log2 (foldchange) and y-axis was −log10 (P value) in volcano plots. The red dots indicate upregulated genes, and green dots indicate downregulated genes. Figure 3. Heatmaps of the shared DEGs in the 3 datasets. The expression of the 195 shared DEGs among KD and controls in each dataset are shown via heatmap. The x-axis represents different samples, and y-axis represents different genes. The red boxes indicate upregulated genes, and blue boxes indicate downregulated genes.
Figure 3. Heatmaps of the shared DEGs in the 3 datasets. The expression of the 195 shared DEGs among KD and controls in each dataset are shown via heatmap. The x-axis represents different samples, and y-axis represents different genes. The red boxes indicate upregulated genes, and blue boxes indicate downregulated genes. Figure 4. GO and KEGG enrichment analyses of shared DEGs. GO enrichment analyses of top 10 most significantly enriched biological process (A), cellular component (B), and molecular function (C) of the shared DEGs, and the top 10 enriched KEGG pathways of the shared DEGs (D).
Figure 4. GO and KEGG enrichment analyses of shared DEGs. GO enrichment analyses of top 10 most significantly enriched biological process (A), cellular component (B), and molecular function (C) of the shared DEGs, and the top 10 enriched KEGG pathways of the shared DEGs (D). Figure 5. PPI network and the most significant module. Upregulated genes are marked in red and downregulated genes are marked in blue in the PPI network (A). The most significant module in PPI network was also identified (B).
Figure 5. PPI network and the most significant module. Upregulated genes are marked in red and downregulated genes are marked in blue in the PPI network (A). The most significant module in PPI network was also identified (B). Figure 6. Predicted miRNA-target DEGs regulatory network. Regulatory network between predicted miRNA and target DEGs. The red dots indicate DEGs and yellow dots indicate miRNAs. The blue lines indicate miRNA-DEGs pairs.
Figure 6. Predicted miRNA-target DEGs regulatory network. Regulatory network between predicted miRNA and target DEGs. The red dots indicate DEGs and yellow dots indicate miRNAs. The blue lines indicate miRNA-DEGs pairs. Figure 7. Immune cell infiltration analysis of the shared DEGs showing the infiltration levels of 22 immune cell subtypes in KD and controls. (A) Stacked bar chart showing deviations of immune infiltration in each control or KD sample among the 3 datasets. (B) Violin plot showing differences in proportions of each immune cell type between control and KD groups. * P<0.05, ** P<0.01, *** P<0.001, ns: not significant.
Figure 7. Immune cell infiltration analysis of the shared DEGs showing the infiltration levels of 22 immune cell subtypes in KD and controls. (A) Stacked bar chart showing deviations of immune infiltration in each control or KD sample among the 3 datasets. (B) Violin plot showing differences in proportions of each immune cell type between control and KD groups. * P<0.05, ** P<0.01, *** P<0.001, ns: not significant. Figure 8. Validation of the hub gene expression changes in KD before and after IVIG treatment. Expression of the identified 10 hub genes in pre- and post-IVIG treatment KD were compared using GSE16797, and 6 genes were significantly downregulated after IVIG. (A) ITGAX. (B) LILRB2. (C) C3AR1. (D) MAPK14. (E) TLR5. (F) MYD88. The x-axis shows different groups and y-axis shows a log2 transformation of gene expression. * P<0.05, ** P<0.01.
Figure 8. Validation of the hub gene expression changes in KD before and after IVIG treatment. Expression of the identified 10 hub genes in pre- and post-IVIG treatment KD were compared using GSE16797, and 6 genes were significantly downregulated after IVIG. (A) ITGAX. (B) LILRB2. (C) C3AR1. (D) MAPK14. (E) TLR5. (F) MYD88. The x-axis shows different groups and y-axis shows a log2 transformation of gene expression. * P<0.05, ** P<0.01. Tables
 Table 1. Gene expression datasets used in this study.
Table 1. Gene expression datasets used in this study. Table 2. The hub genes in Kawasaki disease.
Table 2. The hub genes in Kawasaki disease. Table 1. Gene expression datasets used in this study.
Table 1. Gene expression datasets used in this study. Table 2. The hub genes in Kawasaki disease.
Table 2. The hub genes in Kawasaki disease. Supplementary Table 1. Screening of 195 shared DEGs in KD.
Supplementary Table 1. Screening of 195 shared DEGs in KD. Supplementary Table 2. The top 10 results of GO-BP enrichment analyses of the shared DEGs.
Supplementary Table 2. The top 10 results of GO-BP enrichment analyses of the shared DEGs. Supplementary Table 3. The top 10 results of KEGG pathway analyses of the shared DEGs.
Supplementary Table 3. The top 10 results of KEGG pathway analyses of the shared DEGs. In Press
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
08 Mar 2024 : Laboratory Research
Evaluation of Retentive Strength of 50 Endodontically-Treated Single-Rooted Mandibular Second Premolars Res...Med Sci Monit In Press; DOI: 10.12659/MSM.944110
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








