23 March 2021: Database Analysis
Construction of Decision Trees Based on Gene Expression Omnibus Data to Classify Bladder Cancer and Its Subtypes
Jia-Quan Zhou1C, Xin-Li Kang1DE, Cong-Jie Xu1EF, Shuan Liu1BD, Yang Wang1AG*DOI: 10.12659/MSM.929394
Med Sci Monit 2021; 27:e929394
Abstract
BACKGROUND: Bladder cancer is a malignant tumor of the genitourinary system. Different subtypes of bladder cancer have different treatment methods and prognoses. Therefore, identifying hub genes affecting other genes is of great significance for the treatment of bladder cancer.
MATERIAL AND METHODS: We obtained expression profiles from the GSE13507 and GSE77952 datasets from the Gene Expression Omnibus database. First, principal component analysis was used to identify the difference in gene expression in different types of tissues. Differential expression analysis was used to find the differentially expressed genes between normal and tumor tissues, and between tumors with and without muscle infiltration. Further, based on differentially expressed genes, we constructed 2 decision trees for differentiating between tumor and normal tissues, and between muscle-infiltrating and non-muscle-infiltrating tumor tissues. A receiver operating characteristic curve was used to evaluate the prediction effect of the decision trees.
RESULTS: FAM107A and C8orf4 showed significantly lower expression in bladder cancer tissues than in normal tissues. Regarding muscle infiltration, CTHRC1 showed lower expression and HMGCS2 showed higher expression in non-muscle-infiltrating samples than in those with muscle infiltration. We constructed 2 decision trees for differentiating between tumor and normal tissue, and between tissues with and without muscle infiltration. Both decision trees showed good prediction results.
CONCLUSIONS: These newly discovered hub genes will be helpful in understanding the occurrence and development of different subtypes of bladder cancer, and will provide new therapeutic targets and biomarkers for bladder cancer.
Keywords: Biological Markers, Decision Trees, Biomarkers, Tumor, Databases, Genetic, Extracellular Matrix Proteins, Gene Expression Profiling, Genes, Tumor Suppressor, Hydroxymethylglutaryl-CoA Synthase, Neoplasm Proteins, Nuclear Proteins, Principal Component Analysis, ROC Curve
Background
Recent cancer statistics show that bladder cancer is the second most common malignant tumor among all urogenital tumors [1]. It has become the ninth most common malignant tumor globally and the thirteenth most frequent cause of cancer-related deaths [2]. It is estimated that there are more than 500 000 confirmed cases of bladder cancer each year, and about 200 000 deaths, accounting for 5% of all cancer-related deaths [3]. Currently, the treatment strategies used for bladder cancer usually combine surgery with various adjuvant therapies (such as chemotherapy) [4]. However, for patients with locally advanced or metastatic bladder cancer, the survival rate is still low [4], and the risk of recurrence is high. According to statistics, of the approximately 75% of patients diagnosed with non-muscle-invasive bladder cancer, 30–70% of tumors will reoccur [5], and 30% of tumors will develop into muscle-invasive disease [6]. Therefore, finding new diagnostic and therapeutic targets is important for reducing the mortality of bladder cancer.
Principal component analysis (PCA) is a multivariate statistical method that can identify patterns and classify factors that affect a given phenomenon. It is a technique widely used to identify patterns in the medical field. Machine learning can replace statistical methods, including differential expression [7]. Machine learning algorithms are divided into supervised (with prior knowledge) and unsupervised (without any input of prior knowledge). In the latter, the dataset is divided into training and test data, where the training data are used to create a decision tree and test its performance.
In this study, we first obtained the expression profile for 2 datasets, GSE13507 and GSE77952, from the Gene Expression Omnibus (GEO) database. Evaluation and comparison of gene expression in different kinds of tissues, using PCA, was conducted on the GSE13507 and GSE77952 datasets.
It was determined that the normal and adjacent groups had evident clustering compared with the tumor group samples, and the muscle-infiltrating tumor group and non-muscle-infiltrating tumor group had an obvious difference in clustering. Then, we drew a heatmap based on the gene expression level of the GSE13507 dataset and used a volcano map to show the differentially expressed genes (DEGs) between the normal group and the tumor group in GSE13507. Simultaneously, we draw heatmaps and volcano maps from the GSE77952 dataset to show the DEGs between the muscle-infiltrating tumor group and non-muscle-infiltrating tumor group samples. In the GSE13507 dataset, gene set enrichment analysis (GSEA) and gene ontology (GO) analysis were performed to show biological pathways that may be involved in these different subtypes. Finally, we added the GSE37815, GSE7476, and GSE120736 datasets. After removing batch effects from the expression profiles, samples were randomly divided into a training group and testing group according to the ratio of 7: 3. The decision tree was constructed based on the DEGs from the GSE13507 dataset (normal vs tumor samples). Finally, the expression of the family with sequence similarity 107 member A (FAM107A) and chromosome 8 open reading frame 4 (C8orf4) DEGs was used to classify normal samples vs cancer samples. The differential expression analysis results between the muscle-infiltrating tumor group and the non-muscle-infiltrating tumor group in the GSE13507 dataset and the GSE77952 dataset were selected, and, eventually, 11 DEGs were obtained. Based on these 11 genes, a decision tree classifying samples into either the muscle-infiltrating tumor group or the non-muscle-infiltrating tumor group was constructed. The 2 decision nodes of this classifier were high expression of collagen triple helix repeat containing 1 (CTHRC1) and 3-hydroxymethylglutaryl-CoA synthase 2 (HMGCS2).
This research will help in the understanding of molecular mechanisms underlying bladder cancer and the exploration of muscle-infiltrating and non-muscle-infiltrating tumor types, thereby providing valuable clues for further study.
Material and Methods
DATA COLLECTION:
The GEO database (https://www.ncbi.nlm.nih.gov/geo/) contains high-throughput gene expression and microarray data [8] based on the “GEOquery” R package [9]. We downloaded the expression profiles of 5 datasets: GSE13507 (232 samples), GSE37815 (23 samples), GSE77952 (30 samples), GSE7476 (12 samples), and GSE120736 (112 samples). In total, 124 muscle-invasive samples, 210 non-muscle-invasive samples, and 75 normal samples were included in the study.
PRINCIPAL COMPONENT ANALYSIS:
PCA is a common sample-clustering method usually used for gene expression, diversity analysis, resequencing, and other sample-clustering based on various variable information. We first performed PCA to show clustering of gene expression by sample type, with comparison between tumor, normal, and paracancer tissues in the GSE13507 dataset, comparison between normal, muscle-infiltrating tumor, and non-muscle-infiltrating tumor samples in the GSE13507 dataset, and comparison between muscle-infiltrating tumor and non-muscle-infiltrating tumor samples in the GSE77952 dataset.
DIFFERENTIAL ANALYSIS:
A common differential analysis method for gene expression microarray data is the “Limma” R package [10]. To discover underlying hub genes related to tumorigenesis and muscle infiltration, we performed the following differential analysis: (1) between tumor tissues and normal tissues (normal tissues and pan-cancer tissues) in the GSE13507 dataset, with gene inclusion criteria of P value <0.01 and |logFC| >1.5; (2) between muscle-infiltrating tumor and non-muscle-infiltrating tumor samples in the GSE13507 dataset, with gene inclusion criteria of P value <0.01 and |logFC| >1.2; and (3) between muscle-infiltrating tumor and non-muscle-infiltrating tumor samples in the GSE77952 dataset, with gene inclusion criteria of P value <0.01 and |logFC| >0.4. Volcanic plots were used to show DEGs and their inclusion criteria; heatmaps were used to show the top 100 DEGs.
GENE SET ENRICHMENT ANALYSIS AND ENRICHMENT ANALYSIS:
To explore the potential molecular mechanisms behind our constructed DEGs, enriched terms were found by performing GSEA [11,12]. We utilized the R package “clusterprofiler” [13] to perform GSEA and enrichment analysis. GO terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with adjusted P<0.05 were considered statistically significant and visualized by “GOplot” (R package) [14].
REMOVAL OF BATCH EFFECT:
Since the 5 datasets included in the study were not sequenced from the same batch, further removal of batch effects was needed. Batch effects are technical, systematic biases introduced by sequencing samples that were not correlated with biological status when processed and measured in different batches. In this study, the ComBat function of the “sva” R package was used to remove batch effects and the differences before and after batch effect removal were assessed using PCA.
CONSTRUCTION OF DECISION TREES:
Supervised classification was performed using Recursive Partitioning and Regression Tree (RPART) and was implemented through the “Rpart” R package. After integrating all samples from the 5 datasets, they were randomly divided into training and testing groups (training group: testing group ratio=7: 3). A decision tree was constructed based on the DEGs between normal tissues and tumor tissues in the GSE3507 dataset. The decision tree construction from the muscle-infiltrating tumor and the non-muscle-infiltrating tumor samples was based on the intersection of differentially expressed hub genes in muscle-infiltrating tumor vs non-muscle-infiltrating tumor samples in the GSE13507 dataset and GSE77952 dataset. Receiver operating characteristic curves (ROC curves) were used to evaluate the predictive effect of decision trees in the training and testing sets.
INTERGROUP DIFFERENCES IN HUB MRNAS:
To compare the differences in expression of hub mRNAs in different types of samples, our study compared hub mRNA expression differences between normal and tumor samples in 5 datasets, and the hub mRNA expression differences between muscle-infiltrating tumor and non-muscle-infiltrating tumor samples, using the Wilcoxon test.
Results
PRINCIPAL COMPONENT ANALYSIS:
We performed PCA on the normal samples and tumor samples in the GSE13507 dataset. The results showed significant differences between normal samples (including normal samples and adjacent samples) and tumor samples in the GSE13507 dataset (Figure 1A). Tumor samples and normal samples formed distinct clusters. The PCA of muscle-infiltrated samples and non-muscle-infiltrated samples in the GSE77952 dataset and GSE13507 dataset showed a few differences in clustering between muscle-infiltrating tumors and non-muscle-infiltrating tumors (Figure 1B, 1C).
DIFFERENTIAL EXPRESSION ANALYSIS:
In the GSE13507 dataset, 67 normal samples (including para-cancerous samples) and 165 tumor samples were included. The difference heatmap and volcano map of the 2 groups were drawn according to their different gene expression levels (Figure 2A). Five upregulated genes and 141 downregulated genes emerged from the screening (P value <0.01 and |logFC| >1.5) (Figure 2D). The GSE13507 dataset’s cancer samples included 62 samples that were invasive and 103 samples that were non-invasive. A heatmap of the 2 groups’ gene expression levels was drawn, and a volcano map was used to visualize the DEGs (Figure 2B, 2E). There were 39 upregulated genes and 26 downregulated genes (P value <0.01) and |logFC | >1.2). Finally, in the GSE77952 dataset, we drew a heatmap of between-samples gene expression differences in the muscle-infiltration group vs the non-muscle-infiltration group (Figure 2C), and used the volcano map to show the DEGs (Figure 2F). The screening yielded 45 upregulated and 17 downregulated genes (P value <0.01 and |logFC| >0.4).
GENE SET ENRICHMENT ANALYSIS AND ENRICHMENT ANALYSIS:
To explore the potential mechanism behind the DEGs, GSEA and enrichment analysis were performed. We found that the differential gene enrichment results of the tumor and normal samples were: aminoacyl tRNA biosynthesis, base excision repair, cell cycle, DNA replication, Fanconi anemia pathway, and homologous recombination. In the muscle-infiltrating tumor group and the non-muscle-infiltrating tumor group, the differentially expressed gene sets were enriched in: complement and coagulation cascades, DNA replication, IL-17 signaling pathway, pertussis, prion diseases, and rheumatoid arthritis (Figure 3A, 3B). The GO enrichment analysis showed that the GO annotations of DEGs could be divided into 3 categories: biological processes, cell composition, and molecular functions. We found that DEGs between normal samples and tumor samples were enriched in: antigen processing and presentation of exogenous peptide antigen via major histocompatibility complex (MHC) class II, antigen processing and presentation of peptide antigen via MHC class II, antigen processing and presentation of peptide or polysaccharide antigen via MHC class II, muscle system process, extracellular matrix organization, extracellular structure organization, interferon-gamma-mediated signaling pathway, T-cell receptor signaling pathway, muscle cell differentiation, and enrichment in myofibril assembly (Figure 3C). The DEGs between the infiltrating group and the non-infiltrating group were mainly enriched in: extracellular matrix organization, extracellular structure organization, muscle contraction, cellular zinc ion homeostasis, zinc ion homeostasis, prostate gland morphogenesis, prostate gland development, response to the metal ion, connective tissue development, and detoxification of copper ion (Figure 3D).
REMOVAL OF BATCH EFFECTS:
Before removing batch effects, the direct batch effect of the 5 datasets was first evaluated using PCA, and the results of the analysis are shown in Figure 4A, where the 5 datasets showed separate clustering with obvious differences. The results of the PCA after removing the batch effect through the ComBat function are shown in Figure 4B. There was no separate clustering of expression between the datasets.
CONSTRUCTION OF THE DECISION TREES:
Due to the small sample size of GSE13507 and GSE77952, we added 3 more datasets (GSE37815, GSE7476, and GSE120736). The samples included muscle-invasive tumor (124 cases), non-muscle-invasive tumor (210 cases), and normal tissue (75 cases). The patients were randomly separated into a training set (n=286) and testing set (n=123).
After the grouping was completed, applying RPART to the training set, a decision tree to distinguish normal from tumor tissue was constructed. The decision tree contains 2 decision nodes: FAM107A and C8orf4 (Figure 5A). The testing dataset (30% of the total data) was used to measure the performance of the decision tree. The discriminative power of this classifier was then evaluated by ROC curve. The discriminative power of this decision tree in training and testing sets is shown in Figure 5C, 5D. The area under the curve (AUC) was 0.845 in the training set and 0.7133 in the testing set. That is, the decision tree consisting of FAM107A and C8orf4 showed a good ability to discriminate tumor from normal samples in both the training set and the testing set.
After excluding the normal samples from the training and testing sets, the same method was used for the construction of a decision tree to distinguish between muscle-infiltrating tumor samples and non-muscle-infiltrating tumor samples. This decision tree contained 2 decision nodes, HMGCS2 and CTHRC1 (Figure 5B). The discriminative power of this decision tree in training and testing sets is shown in Figure 5E, 5F. The AUC was 0.7133 in the training set and 0.7038 in the testing set. This indicates that the decision tree, consisting of HMGCS2 and CTHRC1, showed a good ability to discriminate muscle-infiltrating tumor from non-muscle-infiltrating tumor samples in both the training set and the testing set.
INTERGROUP DIFFERENCES IN HUB MRNAS:
To compare the expression differences of FAM107A and C8orf4 in normal and tumor samples, differential expression of HMGCS2 and CTHRC1 was compared in muscle-infiltrating tumor and non-muscle-infiltrating tumor samples. In our study, the expression differences between genes in the 5 datasets were compared using the Wilcoxon test. The results show that FAM107A and C8orf4 both show statistically significantly higher expression in normal samples in the GSE13507, GSE37815, and GSE7476 datasets. GSE77952 and GSE120736 do not include normal tissue samples, so they were not part of this comparison. Compared with the muscle-infiltrating tumor samples, CTHRC1 showed low expression and HMGCS2 showed statistically significantly higher expression in non-muscle-infiltrating tumor samples. This indicates that high expression of FAM107A and C8orf4 may play an important role in the development of bladder cancer, and could possibly be used as a new biomarker for identifying tumors. In tumor samples, HMGCS2 and CTHRC1 might be used to discriminate between muscle-infiltrating tumor and non-muscle-infiltrating tumor samples. Also, the change in expression of HMGCS2 and CTHRC1 may play a large role in the progression from non-muscle-infiltrating to muscle-infiltrating tumors (Figure 6).
Discussion
Bladder cancer is a severe health problem worldwide and is the second most common malignant tumor among all urogenital tumors [1]. Approximately 75% of patients are diagnosed with non-muscle-invasive bladder cancer, and 30–70% of these tumors will reoccur [5]. Unfortunately, the treatment of bladder cancer has made little progress. At present, transurethral bladder tumor resection is the most common surgical method for non-invasive bladder cancer, but the recurrence rate is high [15]. Therefore, there is an urgent need to find new treatment strategies and biomarkers.
In our study, we found that the decision tree constructed based on FAM107A and C8orf4 can be used to distinguish between normal and bladder cancer tissues, as expression of FAM107A and C8orf4 in bladder cancer tissues is reduced compared with that in normal bladder tissues. This finding heralds the potential role of FAM107A and C8orf4 in bladder cancer diagnosis, prognosis, and anti-cancer therapy.
Low expression of FAM107A, in fact, is not only associated with bladder cancer, but may also play an important role in other tumors. A study by Kiwerska et al showed that FAM107A has a low expression level in larynx squamous cell carcinoma [16]. There is also loss of FAM107A expression in non-small cell lung cancer samples, and the key silencing mechanism is not related to promoter hypermethylation [17]. As an activator of the Wnt signaling pathway, C8orf4 is involved in the development of many tumors; for example, cervical squamous cell carcinoma, and high-grade squamous intraepithelial lesions showed significant differences in C8orf4 expression compared with normal cervical tissues or low-grade squamous intraepithelial lesions [18]. Yi-Wen Zheng et al investigated the role of C8orf4 in lung cancers from the perspective of methylation and expression level, and the results showed that the methylation level of C8orf4 in lung cancer tissues was lower than that in normal tissues, and high expression of C8orf4 correlated with poor prognosis [19]. Zhu et al found that C8orf4 showed low expression in liver cancer stem cells and hepatocellular carcinoma tissues, and that self-renewal of liver cancer stem cells is regulated by C8orf4 via suppression of NOTCH2 signaling [20]. These studies all demonstrate the important role of FAM107A and C8orf4 in tumor formation and progression.
Our research also found that HMGCS2 and CTHRC1 can be used to distinguish between muscle-infiltration samples and non-muscle-infiltration samples. In non-muscle-infiltration samples, CTHRC1 showed low expression and HMGCS2 showed high expression. This might mean that CTHRC1 may promote tumor progression. This viewpoint has been supported by gastric cancer studies, in which CTHRC1 was found to be capable of promoting gastric cancer metastasis via the HIF-1α/CXCR4 signaling pathway [21]. In addition, high expression of CTHRC1 has been demonstrated to be closely linked to the prognosis of prostate cancer; immune function related to prostate cancer may be suppressed by CTHRC1; and high expression of CTHRC1 is related to tumor recurrence [22]. CTHRC1 has also been reported to be involved in the progression of liver fibrosis, hepatocellular carcinoma, cervical squamous cell carcinoma, non-small cell lung cancer, and other tumors [23–26]. Similarly, the protective value of HMGCS2 for tumor prognosis has also been demonstrated: HMGCS2 overexpression increased intracellular ketone levels and inhibited cell proliferation, cell migration, and xenograft tumorigenesis in hepatocellular carcinoma [27]. In prostate cancer, HMGCS2 has been shown to act as a tumor suppressor [28]. Tumor inhibition was also demonstrated in esophageal squamous cell carcinoma [29].
Decision trees have been used quite extensively in medicine; for example, decision trees for the selection of surgical approach for hepatectomy for hepatocellular carcinoma [30] and MRI-based decision trees in the diagnosis of biliary atresia in jaundiced infants [31]. Sherafatian et al constructed a decision tree for lung cancer diagnosis and subtype determination based on miRNA expression data in the database [32]. With the development of genomics and the cost reduction of second-generation sequencing, more and more sequencing data are available for our further study, and combining genomics data and decision trees is prudent for cancer research.
In this study, we constructed 2 decision trees for differentiating between tumor and normal tissue and between muscle-infiltrating and non-muscle-infiltrating tumor tissue. These trees will be beneficial for early diagnosis in bladder cancer patients, and may even have the potential to replace traditional diagnostic methods if supported by further studies, which would serve to provide a simple and accurate strategy for the diagnosis of bladder cancer. Meanwhile, the results regarding FAM107A, C8orf4, HMGCS2, and CTHRC1 also suggest important roles for these genes in bladder cancer progression; they may serve as potential therapeutic targets and deserve further investigation.
Conclusions
This study found that FAM107A and C8orf4 show low expression in bladder cancer tissues compared with normal bladder tissues. Also, with regard to muscle-infiltration, CTHRC1 showed lower expression and HMGCS2 showed higher expression in non-muscle-infiltrating tumor samples than in muscle-infiltrating tumor samples. On the bases of our findings, we constructed 2 decision trees for differentiating bladder cancer tumor tissue from normal bladder tissue, and muscle-infiltrating tumor tissue from non-muscle-infiltrating tumor tissue. Both of these decision trees showed good predictive results.
Figures
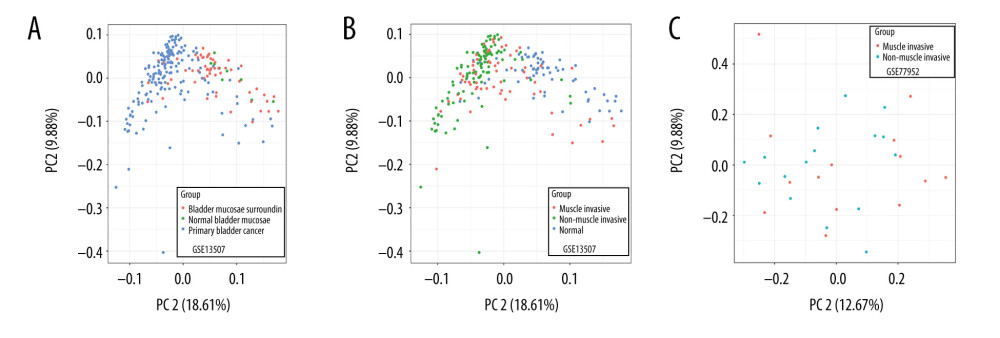 Figure 1. (A–C) Principal component analysis of different types of tissues in the GSE13507 and GSE77952 datasets.
Figure 1. (A–C) Principal component analysis of different types of tissues in the GSE13507 and GSE77952 datasets. 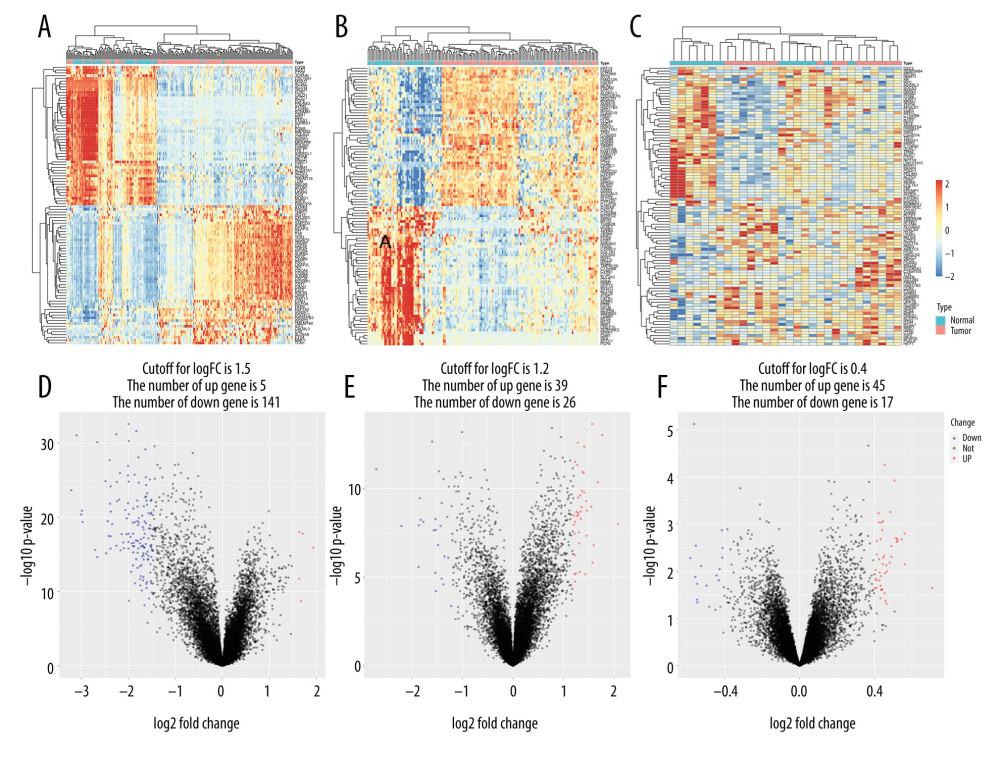 Figure 2. Heatmap and volcano map of different expression genes. (A, D) Normal vs tumor tissue (GSE13507); (B, E) Non-muscle-infiltrating vs muscle-infiltrating tumor tissue (GSE13507); (C, F) Non-muscle-infiltrating vs muscle-infiltrating tumor tissue (GSE77952).
Figure 2. Heatmap and volcano map of different expression genes. (A, D) Normal vs tumor tissue (GSE13507); (B, E) Non-muscle-infiltrating vs muscle-infiltrating tumor tissue (GSE13507); (C, F) Non-muscle-infiltrating vs muscle-infiltrating tumor tissue (GSE77952). 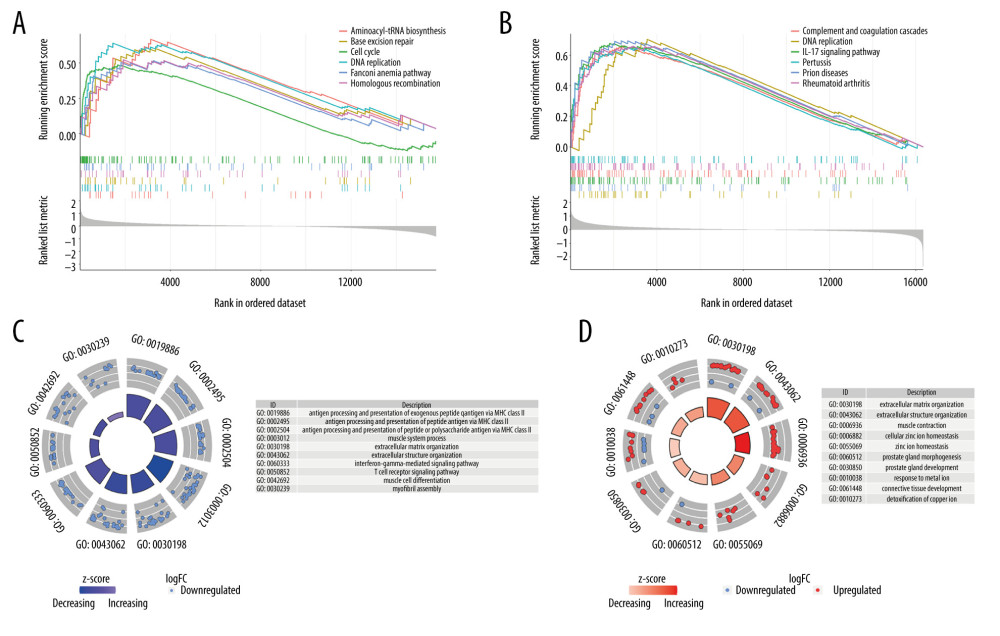 Figure 3. Gene set enrichment analysis and enrichment analysis of differentially expressed genes. (A, C) Differential expression of genes in normal vs tumor tissues in GSE13507. (B, D) Differential expression of genes in non-muscle-infiltrating vs muscle-infiltrating tumors in GSE13507.
Figure 3. Gene set enrichment analysis and enrichment analysis of differentially expressed genes. (A, C) Differential expression of genes in normal vs tumor tissues in GSE13507. (B, D) Differential expression of genes in non-muscle-infiltrating vs muscle-infiltrating tumors in GSE13507. 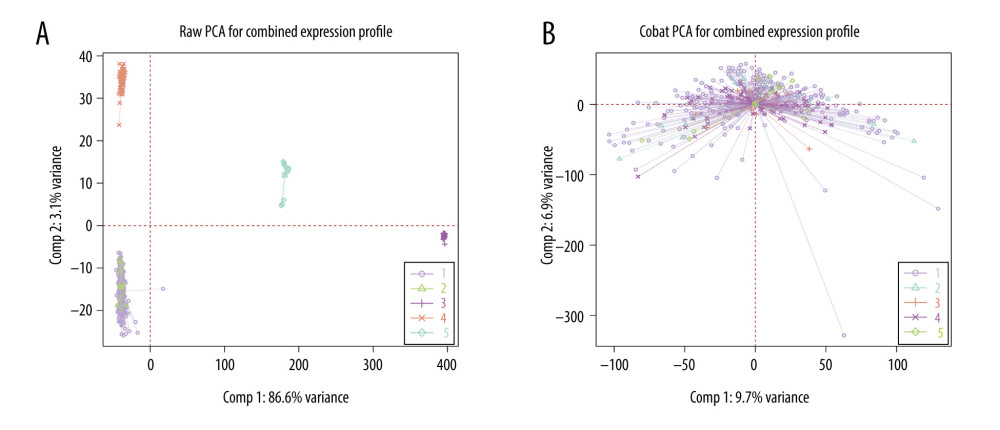 Figure 4. (A, B) Raw principal component analysis (PCA) for combined expression profile and ComBat PCA for combined expression profile.
Figure 4. (A, B) Raw principal component analysis (PCA) for combined expression profile and ComBat PCA for combined expression profile. 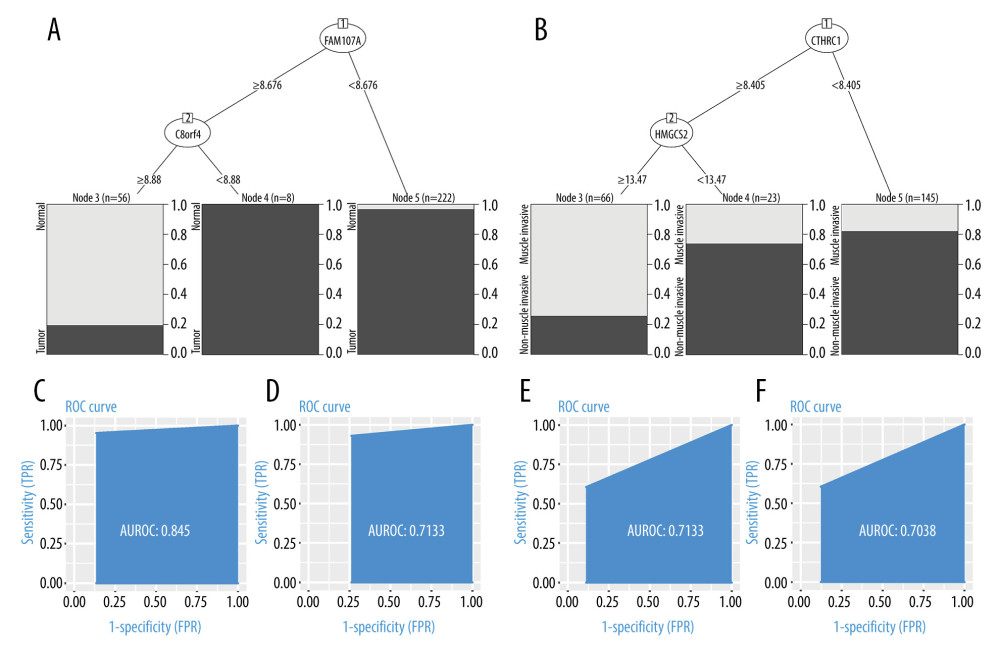 Figure 5. (A, C, D) Decision tree, used to classify normal samples and tumor samples. The area under the curve (AUC) was 0.845 in the training set and 0.7133 in the testing set. (B, E, F) Decision tree, used to classify muscle-infiltrating tumor samples and non-muscle-infiltrating tumor samples, AUC is 0.7133 in the training set and 0.7038 in the testing set.
Figure 5. (A, C, D) Decision tree, used to classify normal samples and tumor samples. The area under the curve (AUC) was 0.845 in the training set and 0.7133 in the testing set. (B, E, F) Decision tree, used to classify muscle-infiltrating tumor samples and non-muscle-infiltrating tumor samples, AUC is 0.7133 in the training set and 0.7038 in the testing set. 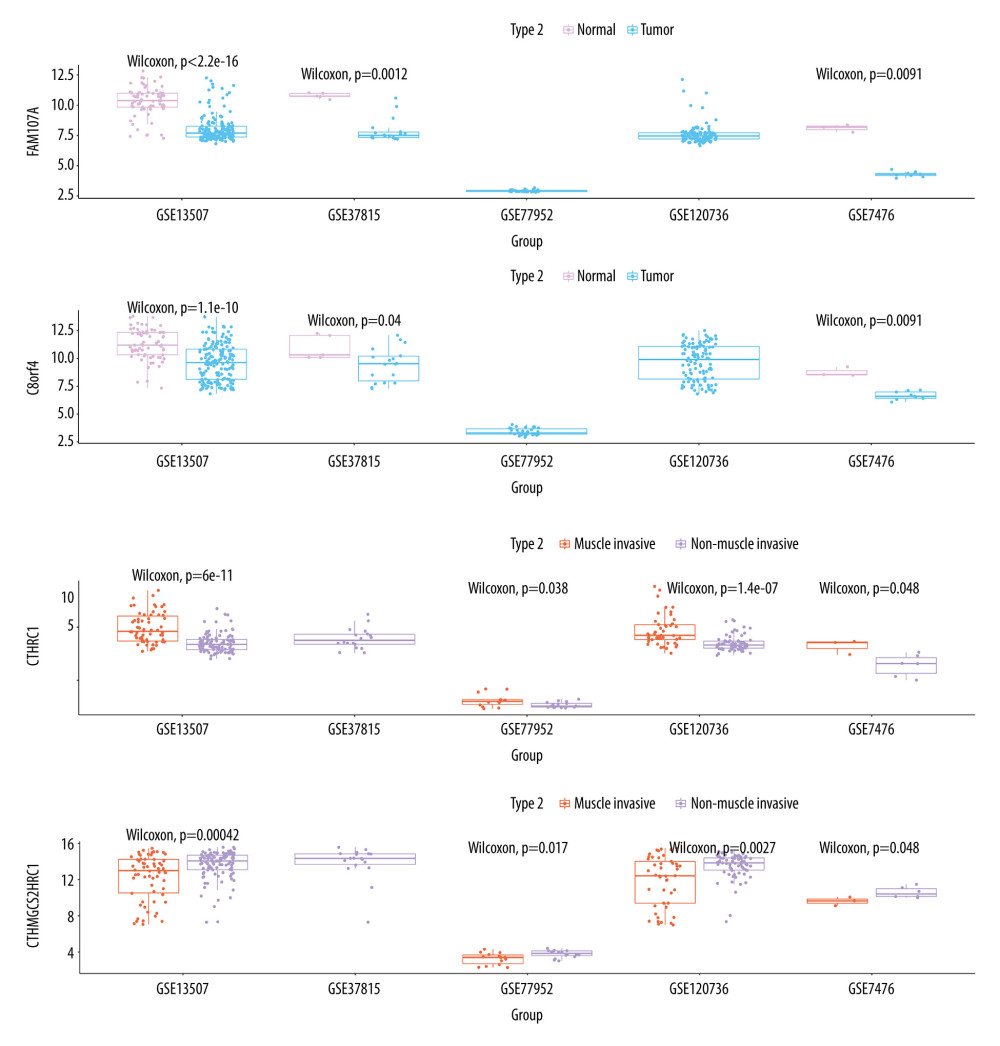 Figure 6. FAM107A and C8orf4 both showed high expression in normal samples from the GSE13507, GSE37815, and GSE7476 datasets. CTHRC1 showed low expression and HMGCS2 showed high expression in the non-muscle-infiltrating tumor samples, compared with the muscle-infiltrating tumor samples.
Figure 6. FAM107A and C8orf4 both showed high expression in normal samples from the GSE13507, GSE37815, and GSE7476 datasets. CTHRC1 showed low expression and HMGCS2 showed high expression in the non-muscle-infiltrating tumor samples, compared with the muscle-infiltrating tumor samples. References
1. Jemal A, Bray F, Center MM, Global cancer statistics: Cancer J Clin, 2011; 61(2); 69-90
2. Ferlay J, Soerjomataram I, Dikshit R, Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012: Int J Cancer, 2015; 136(5); E359-86
3. Bray F, Ferlay J, Soerjomataram I, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries: Cancer J Clin, 2018; 68(6); 394-424
4. Morizane C, Ueno M, Ikeda M, New developments in systemic therapy for advanced biliary tract cancer: Jpn J Clin Oncol, 2018; 48(8); 703-11
5. Babjuk M, Burger M, Zigeuner R, EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2013: Eur Urol, 2013; 64(4); 639-53
6. Millan-Rodriguez F, Chechile-Toniolo G, Salvador-Bayarri J, Primary superficial bladder cancer risk groups according to progression, mortality and recurrence: J Urol, 2000; 164(3 Pt 1); 680-84
7. Geurts P, Irrthum A, Wehenkel L, Supervised learning with decision tree-based methods in computational and systems biology: Mol Biosyst, 2009; 5(12); 1593-605
8. Barrett T, Wilhite SE, Ledoux P, NCBI GEO: Archive for functional genomics data sets – update: Nucleic Acids Res, 2013; 41(Database issue); D991-95
9. Davis S, Meltzer PS, GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor: Bioinformatics, 2007; 23(14); 1846-47
10. Ritchie ME, Phipson B, Wu D, limma powers differential expression analyses for RNA-sequencing and microarray studies: Nucleic Acids Res, 2015; 43(7); e47
11. Mootha VK, Lindgren CM, Eriksson KF, PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes: Nat Genet, 2003; 34(3); 267-73
12. Subramanian A, Tamayo P, Mootha VK, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles: Proc Natl Acad Sci USA, 2005; 102(43); 15545-50
13. Yu G, Wang LG, Han Y, He QY, clusterProfiler: An R package for comparing biological themes among gene clusters: OMICS, 2012; 16(5); 284-87
14. Walter W, Sanchez-Cabo F, Ricote M, GOplot: An R package for visually combining expression data with functional analysis: Bioinformatics, 2015; 31(17); 2912-14
15. Lightfoot AJ, Breyer BN, Rosevear HM, Multi-institutional analysis of sequential intravesical gemcitabine and mitomycin C chemotherapy for non-muscle invasive bladder cancer: Urol Oncol, 2014; 32(1); 35.e15-19
16. Kiwerska K, Szaumkessel M, Paczkowska J, Combined deletion and DNA methylation result in silencing of FAM107A gene in laryngeal tumors: Sci Rep, 2017; 7(1); 5386
17. Pastuszak-Lewandoska D, Czarnecka KH, Migdalska-Sek M, Decreased FAM107A expression in patients with non-small cell lung cancer: Adv Exp Med Biol, 2015; 852; 39-48
18. Lan C, Huan DW, Nie XC, Association of C8orf4 expression with its methylation status, aberrant beta-catenin expression, and the development of cervical squamous cell carcinoma: Medicine (Baltimore), 2019; 98(31); e16715
19. Zheng YW, Zhang L, Wang Y, Thyroid cancer 1 (C8orf4) shows high expression, no mutation and reduced methylation level in lung cancers, and its expression correlates with beta-catenin and DNMT1 expression and poor prognosis: Oncotarget, 2017; 8(38); 62880-90
20. Zhu P, Wang Y, Du Y, C8orf4 negatively regulates self-renewal of liver cancer stem cells via suppression of NOTCH2 signalling: Nat Commun, 2015; 6; 7122
21. Ding X, Huang R, Zhong Y, CTHRC1 promotes gastric cancer metastasis via HIF-1alpha/CXCR4 signaling pathway: Biomed Pharmacother, 2020; 123; 109742
22. Zhou Q, Xiong W, Zhou X, CTHRC1 and PD1/PDL1 expression predicts tumor recurrence in prostate cancer: Mol Med Rep, 2019; 20(5); 4244-52
23. He W, Zhang H, Wang Y, CTHRC1 induces non-small cell lung cancer (NSCLC) invasion through upregulating MMP-7/MMP-9: BMC Cancer, 2018; 18(1); 400
24. Xu G, Fan W, Wang F, CTHRC1 as a novel biomarker in the diagnosis of cervical squamous cell carcinoma: Int J Clin Exp Pathol, 2018; 11(2); 847-54
25. Wang Y, Lee M, Yu G, CTHRC1 activates pro-tumorigenic signaling pathways in hepatocellular carcinoma: Oncotarget, 2017; 8(62); 105238-50
26. Li J, Wang Y, Ma M, Autocrine CTHRC1 activates hepatic stellate cells and promotes liver fibrosis by activating TGF-beta signaling: EBioMedicine, 2019; 40; 43-55
27. Wang YH, Liu CL, Chiu WC, HMGCS2 mediates ketone production and regulates the proliferation and metastasis of hepatocellular carcinoma: Cancers (Basel), 2019; 11(12); 1876
28. Wan S, Xi M, Zhao HB, HMGCS2 functions as a tumor suppressor and has a prognostic impact in prostate cancer: Pathol Res Pract, 2019; 215(8); 152464
29. Tang H, Wu Y, Qin Y, Predictive significance of HMGCS2 for prognosis in resected Chinese esophageal squamous cell carcinoma patients: Onco Targets Ther, 2017; 10; 2553-60
30. Garonzik-Wang JM, Majella Doyle MB, Decision tree for liver resection for hepatocellular carcinoma: JAMA Surg, 2016; 151(9); 853-54
31. Kim YH, Kim MJ, Shin HJ, MRI-based decision tree model for diagnosis of biliary atresia: Eur Radiol, 2018; 28(8); 3422-31
32. Sherafatian M, Arjmand F, Decision tree-based classifiers for lung cancer diagnosis and subtyping using TCGA miRNA expression data: Oncol Lett, 2019; 18(2); 2125-31
Figures
 Figure 1. (A–C) Principal component analysis of different types of tissues in the GSE13507 and GSE77952 datasets.
Figure 1. (A–C) Principal component analysis of different types of tissues in the GSE13507 and GSE77952 datasets. Figure 2. Heatmap and volcano map of different expression genes. (A, D) Normal vs tumor tissue (GSE13507); (B, E) Non-muscle-infiltrating vs muscle-infiltrating tumor tissue (GSE13507); (C, F) Non-muscle-infiltrating vs muscle-infiltrating tumor tissue (GSE77952).
Figure 2. Heatmap and volcano map of different expression genes. (A, D) Normal vs tumor tissue (GSE13507); (B, E) Non-muscle-infiltrating vs muscle-infiltrating tumor tissue (GSE13507); (C, F) Non-muscle-infiltrating vs muscle-infiltrating tumor tissue (GSE77952). Figure 3. Gene set enrichment analysis and enrichment analysis of differentially expressed genes. (A, C) Differential expression of genes in normal vs tumor tissues in GSE13507. (B, D) Differential expression of genes in non-muscle-infiltrating vs muscle-infiltrating tumors in GSE13507.
Figure 3. Gene set enrichment analysis and enrichment analysis of differentially expressed genes. (A, C) Differential expression of genes in normal vs tumor tissues in GSE13507. (B, D) Differential expression of genes in non-muscle-infiltrating vs muscle-infiltrating tumors in GSE13507. Figure 4. (A, B) Raw principal component analysis (PCA) for combined expression profile and ComBat PCA for combined expression profile.
Figure 4. (A, B) Raw principal component analysis (PCA) for combined expression profile and ComBat PCA for combined expression profile. Figure 5. (A, C, D) Decision tree, used to classify normal samples and tumor samples. The area under the curve (AUC) was 0.845 in the training set and 0.7133 in the testing set. (B, E, F) Decision tree, used to classify muscle-infiltrating tumor samples and non-muscle-infiltrating tumor samples, AUC is 0.7133 in the training set and 0.7038 in the testing set.
Figure 5. (A, C, D) Decision tree, used to classify normal samples and tumor samples. The area under the curve (AUC) was 0.845 in the training set and 0.7133 in the testing set. (B, E, F) Decision tree, used to classify muscle-infiltrating tumor samples and non-muscle-infiltrating tumor samples, AUC is 0.7133 in the training set and 0.7038 in the testing set. Figure 6. FAM107A and C8orf4 both showed high expression in normal samples from the GSE13507, GSE37815, and GSE7476 datasets. CTHRC1 showed low expression and HMGCS2 showed high expression in the non-muscle-infiltrating tumor samples, compared with the muscle-infiltrating tumor samples.
Figure 6. FAM107A and C8orf4 both showed high expression in normal samples from the GSE13507, GSE37815, and GSE7476 datasets. CTHRC1 showed low expression and HMGCS2 showed high expression in the non-muscle-infiltrating tumor samples, compared with the muscle-infiltrating tumor samples. In Press
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
08 Mar 2024 : Laboratory Research
Evaluation of Retentive Strength of 50 Endodontically-Treated Single-Rooted Mandibular Second Premolars Res...Med Sci Monit In Press; DOI: 10.12659/MSM.944110
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








