05 October 2023: Clinical Research
Clinical Presentation and Co-Detection of Respiratory Pathogens in Children Under 5 Years with Non-COVID-19 Bacterial and Viral Respiratory Tract Infections: A Prospective Study in Białystok, Poland (2021-2022)
Katarzyna ZdanowiczDOI: 10.12659/MSM.941785
Med Sci Monit 2023; 29:e941785
Abstract
BACKGROUND: Respiratory tract infections (RTIs) in children often involve a complex interplay between viruses and bacteria. This study aimed to evaluate clinical presentation in children under 5 years old diagnosed with non-COVID-19 bacterial and viral respiratory tract co-infections between October 2021 and May 2022 in Białystok, Poland.
MATERIAL AND METHODS: We recruited 100 children under 5 years with RTIs who tested negative for SARS-CoV-2. Nasopharyngeal swabs were screened for 19 viruses and 7 bacterial strains using molecular assays.
RESULTS: Viral pathogens were detected in 71% of patients and bacterial pathogens were detected in 59%. The most common pathogens were Haemophilus influenzae (n=48), rhinoviruses (n=32), and Streptococcus pneumoniae (n=30). Single pathogens were detected in 36%, dual in 37%, triple in 15%, and quadruple in 2%. Bacterial pathogens were co-detected with viruses in 40 cases, mostly with rhinoviruses (n=15). Two different viruses were found in 14 children and the most common co-detection was adenovirus with rhinovirus (n=5); dyspnea (63% vs 11%) and wheezing (75% vs 22%) were more common in children with human bocavirus. Fever was a common symptom in children with human adenovirus (88% vs 58%). Detection of bacteria and multiple detections were more common in day-care attendees, but were not associated with clinical picture of RTI.
CONCLUSIONS: Consistent with previous studies, we found a high prevalence of rhinoviruses, despite ongoing implementation of non-pharmaceutical interventions to contain the COVID-19 pandemic. Co-detection of 2 different respiratory pathogens was frequent, but we found no evidence that this was associated with the severity of infections.
Keywords: coinfection, Influenza, Human, Respiratory Syncytial Virus, Human, Respiratory Tract Infections, Streptococcus pneumoniae, Humans, Child, Infant, Child, Preschool, Prospective Studies, Poland, Pandemics, COVID-19, SARS-CoV-2, Viruses, Bacteria, Rhinovirus
Background
Respiratory tract infections (RTIs) cause a wide spectrum of illnesses, from the common cold to severe pneumonia [1]. Understanding the etiology of these infections is important not only in everyday clinical practice, but also in public health. It enables the introduction of prevention methods (including vaccinations and early isolation of specific individuals), and the application of appropriate treatment, reducing the overuse of antibiotics. Most RTIs are caused by viruses, but it is clinically impossible to distinguish them from bacterial infections [2]. The seasonality and etiology of RTI was profoundly altered during the coronavirus disease 2019 (COVID-19) pandemic. Global influenza virus and respiratory syncytial virus (RSV) activity decreased markedly with the introduction of non-pharmaceutical interventions (NPIs) in the COVID-19 pandemic [3]. However, several non-SARS-CoV-2 respiratory pathogens were still circulating in communities [4]. Despite the ongoing implementation of community NPIs, the prevalence of rhinoviruses and respiratory enteroviruses rebounded rapidly and persisted [4]. Evidence from the initial wave of the COVID-19 pandemic in early 2020 suggests that co-infections of SARS-CoV-2 with other respiratory viruses were also observed. The incidence of these co-infections varied significantly, and their impact on clinical severity and prognosis remains unclear [5]. These findings highlight the ongoing prevalence and impact of other respiratory viruses during the pandemic, emphasizing the need for comprehensive surveillance and understanding of RTIs beyond COVID-19.
Conventional methods of pathogen detection, such as cultures of nasopharyngeal aspirates or throat swabs, serological titters, and antigen detection, often do not have sufficient sensitivity, are time-consuming, and require extensive equipment and qualified staff, which often makes them inaccessible in everyday practice [2]. The introduction of molecular methods in recent years has significantly improved the detection of pathogens causing infections, especially viruses. Multiplex polymerase chain reaction (PCR) tests simultaneously detect several pathogenic viruses and bacteria in a short time and with high sensitivity, so they can improve patient management [6,7].
Therefore, this study aimed to evaluate outcomes in 100 children under age 5 years diagnosed with non-COVID-19 bacterial and viral lower respiratory tract co-infections between October 2021 and May 2022 in Białystok, Poland.
Material and Methods
ETHICS STATEMENT:
The protocol was approved by the Bioethics Committee of the Medical University of Białystok prior to patient recruitment, and the study was conducted in accordance with the Helsinki Accords (protocol code APK.002.420.2020 approved on 17 December 2020). Informed consent was collected from parents/guardians of all study participants.
STUDY DESIGN:
This was a prospectively recruited observational cohort study done in Białystok, Poland between October 2021 and May 2022. We included 100 children under 5 years of age with symptoms of upper and lower RTI. To address potential sources of bias, the samples were collected from 1 family doctor clinic and 2 pediatric wards in Białystok, Poland. The number of nasopharyngeal swabs (NPSs) taken per week depended on the number of patients with RTI symptoms. To cover a wider time frame, the maximum number of samples collected per week could not exceed 10 NPSs. Clinical and demographic data were collected using a questionnaire completed by the consulting doctor. The NPS were obtained from patients on the day they were included in the study. The criteria for including patients in the study were the presence of signs of acute RTI (2 of the following: fever, runny nose, cough, dyspnea) and a negative test for SARS-CoV-2 infection in the GeneXpert assays (Cepheid; Sunnyvale, CA, USA). Routine blood tests (complete blood count and C-reactive protein) collected from hospitalized patients using standard methods were also recorded. Patients pre-treated with antibiotics, or diagnosed with chronic respiratory diseases or non-infectious diseases were excluded from the analysis.
MOLECULAR DETECTION OF RESPIRATORY PATHOGENS:
NPSs were collected by a standard method according to the instrument capacity and immediately put into a tube containing transport medium. Following the sample collection, nucleic acids were extracted from nasopharyngeal swabs with the use of magnetic bead method (MagPurix EVO 24 CE-IVD Nucleic Acid Extraction System, Zinexts Life Sciences Corp., Taipei, Taiwan). Furthermore, molecular screening for the particular infectious agents was performed utilizing the nucleic acid amplification test (NAAT) with subsequent real-time signal detection performed using the CFX96TM Real-Time PCR Detection System (Bio-Rad, Hercules, California, USA).
Molecular screening covered RNA viruses (HMPV – human metapneumovirus; EV – enterovirus; HPIVs – human parainfluenza viruses 1–4; HRV – human rhinovirus A/B/C; HCoV 229E – human coronavirus 229E; HCoV NL63 – human coronavirus NL63; HCoV OC43 – human coronavirus OC43; FluA – influenza A virus with H1, H3, and H1pdm09 subtyping; FluB – influenza B virus; hRSV-A – human respiratory syncytial virus subtype A; hRSV-B – human respiratory syncytial virus subtype B), DNA viruses (HAdV – human adenovirus; HBoV – human bocavirus 1/2/3/4), and bacteria (
STATISTICAL ANALYSIS:
Statistical analysis was done using TIBCO Software Inc. (2017) Statistica, version 13 (Palo Alto, CA, USA) and GraphPad Prism version 9.4.0 for Windows, GraphPad Software. The quantitative data are expressed as median, maximum, and minimum values. The qualitative variables are expressed as absolute frequency and percentage. Comparative statistics included the chi-square test and Mann-Whitney U test. Multivariable logistic regression models were used to identify variables that were associated with the risk associated with the detection of at least 2 pathogens. In these models the types of pathogens detected (bacterial/viral/viral-viral/bacterial-viral) were modeled against the presence of symptoms and demographic data of patients. Statistical significance was defined as a
Results
CHARACTERISTICS OF THE STUDIED POPULATION:
The median age of patients was 24 months, and 54% of patients were male. Almost half of the patients (45%) were toddlers aged 13–26 months. Among the participants of the study, 56% were hospitalized in pediatric departments and 44% were consulted in the outpatient clinic. Most of the children (57%) were attending day-care units (a nursery or a kindergarten). The demographic features of the studied group are shown in the Table 1.
DETECTION OF RESPIRATORY PATHOGENS IN THE NASOPHARYNGEAL SWABS:
Among 100 respiratory samples tested, 90 were positive and 10 were negative for pathogens. A total of 15 different pathogens were detected, and co-detections were common (54% of all patients). The highest incidence of RTIs was observed from November to January. However, we did not observe any apparent seasonality of detected pathogens (Figure 1). Viral pathogens were detected in 66% of outpatients and 75% of hospitalized children. Bacteria were detected in 59% of hospitalized and 59% of patients consulted in the clinic. Haemophilus influenzae (48%) and Streptococcus pneumoniae (30%) were the only 2 bacterial pathogens detected in this study. Human rhinovirus (32%) was the leading viral pathogen, followed by HAdV (16%) (Table 2). A single detection was seen in 36%, dual in 37%, triple in 15%, and quadruple in 2% of patients. Bacterial pathogens were co-detected with viruses in 40 children. This constitutes 68% (40/59) of patients in whom a bacterial pathogen was detected (Table 3). Rhinoviruses were the most common viruses co-detected with bacterial pathogens (n=15; 37.5%). Two different viruses were found in 14 children and the most common co-detection was adenovirus with rhinovirus (n=5; 36%). The influenza A virus was detected in 2 children (both in March 2022 only) and RSV A/B were identified in 5 patients (between December 2021 and February 2022). The following pathogens were detected in none of the patients: HPIV1, HPIV4, Flu A H1, Flu A H1pmd09, Flu B, Chlamydophila pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila, Bordetella pertussis, and Bordetella parapertussis.
RESPIRATORY PATHOGENS ASSOCIATED WITH CLINICAL SYMPTOMS:
Wheezing (75% vs 22%; risk-ratio [RR] 10.8; 95% confidence interval [CI], 2.0–57.7; p=0.005) and dyspnea (63% vs 11% RR, 13.7; 95% CI, 2.8–66.0; p=0.001) were more likely to be present in infections caused by the HBoV, compared to other etiologies. Infections with HAdV were typically characterized by the presence of fever (88% vs 58%; RR, 5.0; 95% CI 1.1–23.4; p=0.04), but cough was slightly less common (56% vs 80%; RR, 0.3; 95% CI, 0.1–1.0; p=0.05), than in infections caused by other pathogens (Figure 2). The clinical picture of the disease was not associated with the presence of other pathogens, multiple detections, or with the detection of bacterial pathogens, as revealed by regression models. However, we found that children with a total number of symptoms equal to or higher than median (4 symptoms) were more likely to test positive for at least 1 virus (RR, 2.96; 95% CI 1.21–7.20; p=0.015). Detection of 2 different viruses or mixed viral-bacterial detection was not associated with the number of observed symptoms.
FACTORS INFLUENCING DETECTION OF BACTERIAL PATHOGENS AND MULTIPLE DETECTIONS:
The risk of detecting bacterial pathogens was dependent on the time spent in a day-care unit. In the univariate analysis, the RR of detecting S. pneumoniae and H. influenzae in children spending over 40 h weekly at a day-care unit was 3.66 (95% Cl, 1.16–11.56; p=0.027) and 7.11 (95% CI, 2.00–25.24; p=0.002), respectively, compared to children who spent there less than 20 h weekly or did not attend day-care units at all. In the multivariable analysis, we found that longer hours at day-care units was the only significant factor associated with RR=11.02 of detecting any bacterial pathogens in NPSs (95% CI, 1.16–104.36; p=0.035). The risk of multiple detections was also higher in this group of children (Figure 3).
The risk of detecting none of the viruses was associated with attendance at day-care units. In 14 children, 2 different viruses were detected. This co-detection had no impact on the clinical symptoms. None of the analyzed demographic factors increased the risk of viral coinfection.
CLINICAL PRESENTATION IN HOSPITALIZED PATIENTS:
Compared to ambulatory care, children who were hospitalized more often had wheezing (42.9% vs 4.5%;
Concentrations of C-reactive protein (CRP) over 40 mg/L were seen in 13 (23%) of hospitalized patients. Children with elevated concentrations of CRP were more likely to test positive for HAdV (RR, 5.29; 95% CI, 1.32–21.23;
USE OF ANTIBIOTICS:
Antibiotics were used in 45% of patients. Patients who tested positive for HAdV were more likely to receive antibiotics (RR, 4.17; 95% CI, 1.09–15.91;
Discussion
In this study we found that during the COVID-19 pandemic, the detection of non-SARS-CoV-2 respiratory pathogens in RTI children was high and reached 90%, which was similar to the pre-pandemic period [8,9]. In contrast to before the COVID-19 pandemic, when RSV and flu predominated, in this study rhinoviruses were the most commonly detected viruses. Co-detection of 2 different respiratory pathogens was common, but was not associated with clinical presentation of the infection.
RTIs are among the most common reasons for pediatric primary care consultations and hospital admissions, posing a significant health burden [10–12]. Despite greater knowledge about the etiology of infection, in practice it is difficult to unequivocally identify the pathogen responsible for RTI, and thus to apply appropriate antimicrobial treatment. The main pathogens detected in this study were
The most frequent viruses reported in our analysis were HRV and HAdV, which were commonly detected in patients with RTIs before [25,26] and during the COVID-19 pandemic [27,28]. On the other hand, the RSV and influenza viruses were rarely detected in this study compared to the pre-pandemic period [7,29]. We found that RSV and influenza virus were actively circulating in early 2022 in Poland, possibly leading the influenza and RSV outbreaks in late 2022. These outbreaks of non-COVID-19 respiratory infections were observed across the world [30–32]. A decrease in the number of COVID-19 cases was followed by an increase in prevalence of non-COVID-19 pathogens, indicating interactions between respiratory pathogens [34]. A recent study from Austria showed that the surge of RSV infections observed in 2022 was caused by lineages already present before the pandemic [33].
Bacterial-bacterial, bacterial-viral, and viral-viral co-detections were common, but did not add to the severity of disease in this study. Previous studies have suggested that polymicrobial infections enhance the RTI severity in children [35,36]. In contrast to our observations, in these studies the prevalence of RSV and influenza infections was high. Possibly, interactions between RSV and influenza viruses with bacteria in the pediatric respiratory tract contribute to greater disease severity than when other viruses are present.
Our study has several important limitations. First, the sample size was small. A larger group could have allowed for a better understanding of factors affecting detection of respiratory pathogens in RTI children. Second, we did not recruit healthy controls for comparison, so we could not determine whether the detected organisms were the causes of RTI, since those organisms are commonly found in healthy children. Third, this study was done in a single hospital and a single outpatient clinic, so the detected pathogens might not be representative of the true epidemiology. Moreover, although sensitivity of molecular methods used in this study is considered to be high, we cannot rule out the possibility that some of the pathogens were missed because of the limitations of this approach, including inhibition of the amplification reaction or the lack of a specific target [37].
Conclusions
The findings of this study show that rhinoviruses and other non-SARS-CoV-2 viral and bacterial pathogens were commonly detected in children under age 5, despite ongoing implementation of non-pharmaceutical interventions to contain the COVID-19 pandemic. Co-detection of 2 different bacterial and viral respiratory pathogens was common, but did not add to the severity of acute respiratory infections. Possible mechanisms that drive these bacterial-viral interactions require further investigation.
Figures
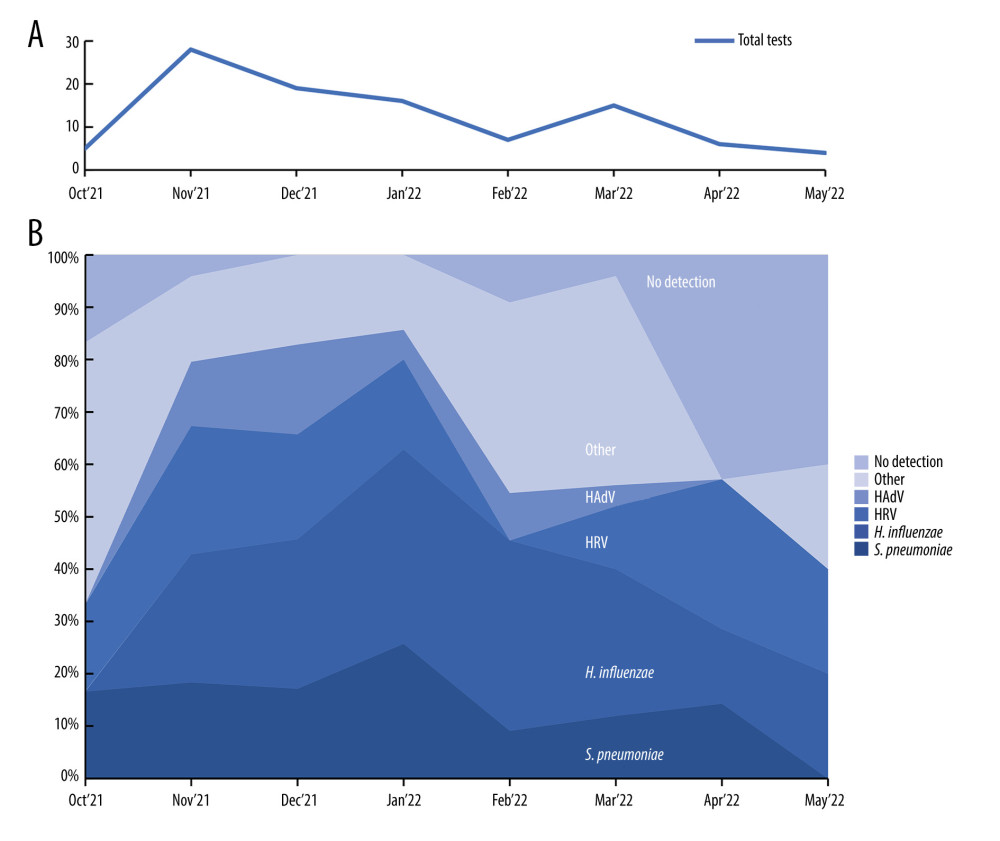 Figure 1. Distribution of respiratory pathogens over the study period. (A) Monthly number of nasopharyngeal swabs collected during the study; (B) Stack area chart showing the most common pathogens detected during the study period. HRV – human rhinovirus; HAdV – human adenovirus. This figure was created using Microsoft Corporation, 2021. Microsoft Excel.
Figure 1. Distribution of respiratory pathogens over the study period. (A) Monthly number of nasopharyngeal swabs collected during the study; (B) Stack area chart showing the most common pathogens detected during the study period. HRV – human rhinovirus; HAdV – human adenovirus. This figure was created using Microsoft Corporation, 2021. Microsoft Excel. 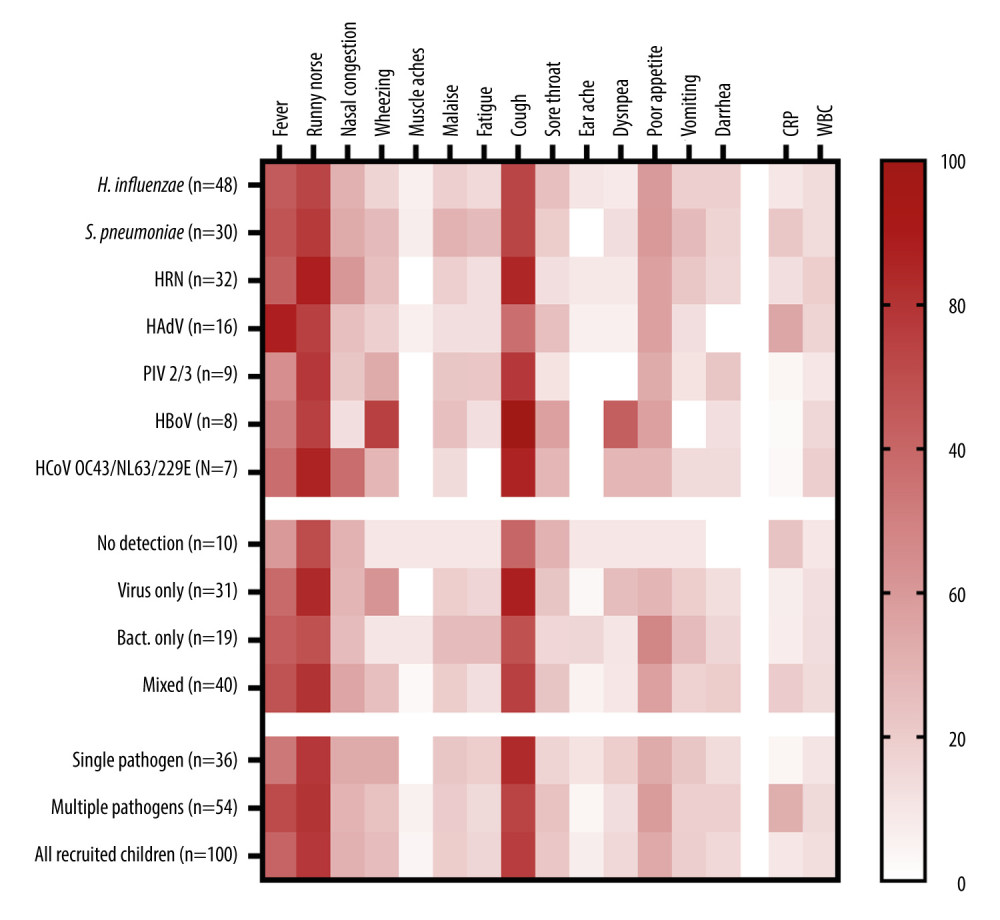 Figure 2. The heatmap of clinical signs and inflammatory markers divided by the type of pathogen detected and the presence of co-detections. Symptoms are shown as percentages, CRP as mg/L, and WBC as 103 cells/μL. CRP and WBC are shown as median values. HRV – human rhinovirus; HAdV – human adenovirus; PIV – parainfluenza virus; HBoV – human bocavirus; HCoV – human coronavirus; CRP – C-reactive protein; WBC – white blood cell count. This figure was created using GraphPad Prism version 10.0.0 for Windows, GraphPad Software, Boston, Massachusetts USA.
Figure 2. The heatmap of clinical signs and inflammatory markers divided by the type of pathogen detected and the presence of co-detections. Symptoms are shown as percentages, CRP as mg/L, and WBC as 103 cells/μL. CRP and WBC are shown as median values. HRV – human rhinovirus; HAdV – human adenovirus; PIV – parainfluenza virus; HBoV – human bocavirus; HCoV – human coronavirus; CRP – C-reactive protein; WBC – white blood cell count. This figure was created using GraphPad Prism version 10.0.0 for Windows, GraphPad Software, Boston, Massachusetts USA. 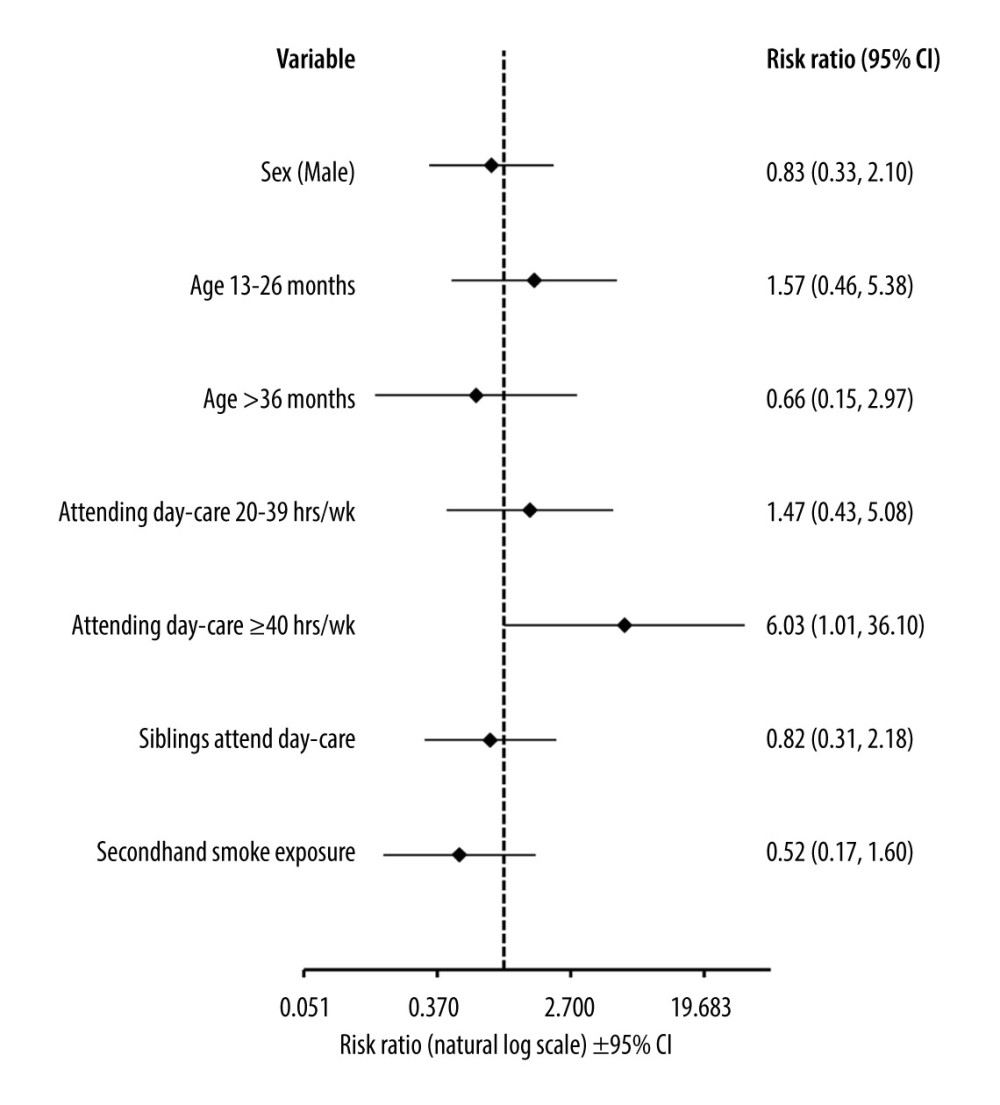 Figure 3. Forest plot graph showing factors associated with detecting more than 1 pathogen in nasopharyngeal swabs. Data calculated using the multivariable logit regression analysis. Age-related risk was calculated with relation to age under 12 months. Risk associated with attending day-care was calculated with relation to spending less than 20 h per week or not attending day-care units at all. CI – confidence interval. This figure was created using Microsoft Corporation, 2021. Microsoft Excel.
Figure 3. Forest plot graph showing factors associated with detecting more than 1 pathogen in nasopharyngeal swabs. Data calculated using the multivariable logit regression analysis. Age-related risk was calculated with relation to age under 12 months. Risk associated with attending day-care was calculated with relation to spending less than 20 h per week or not attending day-care units at all. CI – confidence interval. This figure was created using Microsoft Corporation, 2021. Microsoft Excel. References
1. Simoes EAF, Cherian T, Chow J, Acute respiratory infections in children: Disease Control Priorities in Developing Countries, 2006, Washington (DC), The International Bank for Reconstruction and Development/The World Bank
2. Das S, Dunbar S, Tang Y-W, Laboratory diagnosis of respiratory tract infections in children – the state of the art: Front Microbiol, 2018; 9; 2478
3. Chow EJ, Uyeki TM, Chu HY, The effects of the COVID-19 pandemic on community respiratory virus activity: Nat Rev Microbiol, 2023; 21(3); 195-210
4. Duclos M, Hommel B, Allantaz F, Multiplex PCR detection of respiratory tract infections in SARS-CoV-2-negative patients admitted to the Emergency Department: An international multicenter study during the COVID-19 pandemic: Microbiol Spectr, 2022; 10(5); e02368-22
5. Maltezou HC, Papanikolopoulou A, Vassiliu S, COVID-19 and respiratory virus co-infections: A systematic review of the literature: Viruses, 2023; 15(4); 865
6. Alamri AM, Alkhilaiwi FA, Ullah Khan N, Era of molecular diagnostics techniques before and after the COVID-19 pandemic: Curr Issues Mol Biol, 2022; 44(10); 4769-89
7. Visseaux B, Collin G, Ichou H, Usefulness of multiplex PCR methods and respiratory viruses’ distribution in children below 15 years old according to age, seasons and clinical units in France: A 3 years retrospective study: PLoS One, 2017; 12(2); e0172809
8. Freymuth F, Vabret A, Cuvillon-Nimal D, Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness: J Med Virol, 2006; 78(11); 1498-504
9. Lamrani Hanchi A, Guennouni M, Rachidi M, Epidemiology of respiratory pathogens in children with severe acute respiratory infection and impact of the multiplex PCR film array respiratory panel: A 2-year study: Int J Microbiol, 2021; 2021; 2276261
10. Jin X, Ren J, Li R, Global burden of upper respiratory infections in 204 countries and territories, from 1990 to 2019: eClinicalMedicine, 2021; 37; 100986
11. Li Y, Nair H, Trends in the global burden of lower respiratory infections: The knowns and the unknowns: Lancet Infect Dis, 2022; 22(11); 1523-25
12. Hasegawa K, Tsugawa Y, Cohen A, Camargo CA, Infectious disease-related Emergency Department visits among children in the US: Pediatr Infect Dis J, 2015; 34(7); 681-85
13. Kovács E, Sahin-Tóth J, Tóthpál A: PLoS One, 2020; 15(2); e0229021
14. Hu J, Sun X, Huang Z: BMC Infect Dis, 2016; 16; 149
15. Navne JE, Børresen ML, Slotved HC, Nasopharyngeal bacterial carriage in young children in Greenland: A population at high risk of respiratory infections: Epidemiol Infect, 2016; 144(15); 3226-36
16. Tsai M-H, Liao S-L, Chiu C-Y, Longitudinal investigation of nasopharyngeal pneumococcal carriage in early childhood: The PATCH birth cohort study: PLoS One, 2020; 15(8); e0237871
17. Tikhomirova A, Kidd SP: Pathog Dis, 2013; 69(2); 114-26
18. Haddadin Z, Schuster JE, Spieker AJ, Acute respiratory illnesses in children in the SARS-CoV-2 pandemic: Prospective multicenter study: Pediatrics, 2021; 148(2); e2021051462
19. Fourgeaud J, Toubiana J, Chappuy H, Impact of public health measures on the post-COVID-19 respiratory syncytial virus epidemics in France: Eur J Clin Microbiol Infect Dis, 2021; 40(11); 2389-95
20. Liu P, Xu M, Cao L, Impact of COVID-19 pandemic on the prevalence of respiratory viruses in children with lower respiratory tract infections in China: Virol J, 2021; 18; 159
21. Kishimoto K, Bun S, Shin J-H, Early impact of school closure and social distancing for COVID-19 on the number of inpatients with childhood non-COVID-19 acute infections in Japan: Eur J Pediatr, 2021; 180(9); 2871-78
22. Todd IMF, Miller JE, Rowe SL, Changes in infection-related hospitalizations in children following pandemic restrictions: An interrupted time-series analysis of total population data: Int J Epidemiol, 2021; 50(5); 1435-43
23. Angoulvant F, Ouldali N, Yang DD, Coronavirus disease 2019 pandemic: Impact caused by school closure and national lockdown on pediatric visits and admissions for viral and nonviral infections-a time series analysis: Clin Infect Dis, 2021; 72(2); 319-22
24. Wróbel-Pawelczyk I, Ronkiewicz P, Wanke-Rytt M, Pneumococcal carriage in unvaccinated children at the time of vaccine implementation into the national immunization program in Poland: Sci Rep, 2022; 12(1); 5858
25. Tsagarakis NJ, Sideri A, Makridis P, Age-related prevalence of common upper respiratory pathogens, based on the application of the FilmArray Respiratory panel in a tertiary hospital in Greece: Medicine (Baltimore), 2018; 97(22); e10903
26. Bicer S, Giray T, Çöl D, Virological and clinical characterizations of respiratory infections in hospitalized children: Ital J Pediatr, 2013; 39(1); 22
27. Chiu Y-T, Tien N, Lin H-C, Detection of respiratory pathogens by application of multiplex PCR panel during early period of COVID-19 pandemic in a tertiary hospital in Central Taiwan: J Microbiol Immunol Infect, 2022; 55(6 Pt 2); 1144-50
28. Diesner-Treiber SC, Voitl P, Voitl JJM, Respiratory infections in children during a COVID-19 pandemic winter: Front Pediatr, 2021; 9; 740785
29. Rha B, Curns AT, Lively JY, Respiratory syncytial virus-associated hospitalizations among young children: 2015–2016: Pediatrics, 2020; 146(1); e20193611
30. Ben Moussa M, Buckrell S, Rahal A, National influenza mid-season report, 2022–2023: A rapid and early epidemic onset: Can Commun Dis Rep, 2023; 49(1); 10-14
31. World Health Organization, Global Influenza Programme: Influenza updates https://www.who.int/teams/global-influenza-programme/surveillance-and-monitoring/influenza-updates
32. Riepl A, Straßmayr L, Voitl P, The surge of RSV and other respiratory viruses among children during the second COVID-19 pandemic winter season: Front Pediatr, 2023; 11; 1112150
33. Redlberger-Fritz M, Springer DN, Aberle SW, Respiratory syncytial virus surge in 2022 caused by lineages already present before the COVID-19 pandemic: J Med Virol, 2023; 95(6); e28830
34. Wang Q, Jia M, Jiang M, Seesaw effect between COVID-19 and influenza from 2020 to 2023 in World Health Organization regions: Correlation analysis: JMIR Public Health Surveill, 2023; 9; e44970
35. Brealey JC, Sly PD, Young PR, Chappell KJ, Viral bacterial co-infection of the respiratory tract during early childhood: FEMS Microbiol Lett, 2015; 362(10); fnv062
36. Brealey JC, Chappell KJ, Galbraith S: Respirol Carlton Vic, 2018; 23(2); 220-27
37. García-Arroyo L, Prim N, Martí N, Benefits and drawbacks of molecular techniques for diagnosis of viral respiratory infections. Experience with two multiplex PCR assays: J Med Virol, 2016; 88(1); 45-50
Figures
 Figure 1. Distribution of respiratory pathogens over the study period. (A) Monthly number of nasopharyngeal swabs collected during the study; (B) Stack area chart showing the most common pathogens detected during the study period. HRV – human rhinovirus; HAdV – human adenovirus. This figure was created using Microsoft Corporation, 2021. Microsoft Excel.
Figure 1. Distribution of respiratory pathogens over the study period. (A) Monthly number of nasopharyngeal swabs collected during the study; (B) Stack area chart showing the most common pathogens detected during the study period. HRV – human rhinovirus; HAdV – human adenovirus. This figure was created using Microsoft Corporation, 2021. Microsoft Excel. Figure 2. The heatmap of clinical signs and inflammatory markers divided by the type of pathogen detected and the presence of co-detections. Symptoms are shown as percentages, CRP as mg/L, and WBC as 103 cells/μL. CRP and WBC are shown as median values. HRV – human rhinovirus; HAdV – human adenovirus; PIV – parainfluenza virus; HBoV – human bocavirus; HCoV – human coronavirus; CRP – C-reactive protein; WBC – white blood cell count. This figure was created using GraphPad Prism version 10.0.0 for Windows, GraphPad Software, Boston, Massachusetts USA.
Figure 2. The heatmap of clinical signs and inflammatory markers divided by the type of pathogen detected and the presence of co-detections. Symptoms are shown as percentages, CRP as mg/L, and WBC as 103 cells/μL. CRP and WBC are shown as median values. HRV – human rhinovirus; HAdV – human adenovirus; PIV – parainfluenza virus; HBoV – human bocavirus; HCoV – human coronavirus; CRP – C-reactive protein; WBC – white blood cell count. This figure was created using GraphPad Prism version 10.0.0 for Windows, GraphPad Software, Boston, Massachusetts USA. Figure 3. Forest plot graph showing factors associated with detecting more than 1 pathogen in nasopharyngeal swabs. Data calculated using the multivariable logit regression analysis. Age-related risk was calculated with relation to age under 12 months. Risk associated with attending day-care was calculated with relation to spending less than 20 h per week or not attending day-care units at all. CI – confidence interval. This figure was created using Microsoft Corporation, 2021. Microsoft Excel.
Figure 3. Forest plot graph showing factors associated with detecting more than 1 pathogen in nasopharyngeal swabs. Data calculated using the multivariable logit regression analysis. Age-related risk was calculated with relation to age under 12 months. Risk associated with attending day-care was calculated with relation to spending less than 20 h per week or not attending day-care units at all. CI – confidence interval. This figure was created using Microsoft Corporation, 2021. Microsoft Excel. Tables
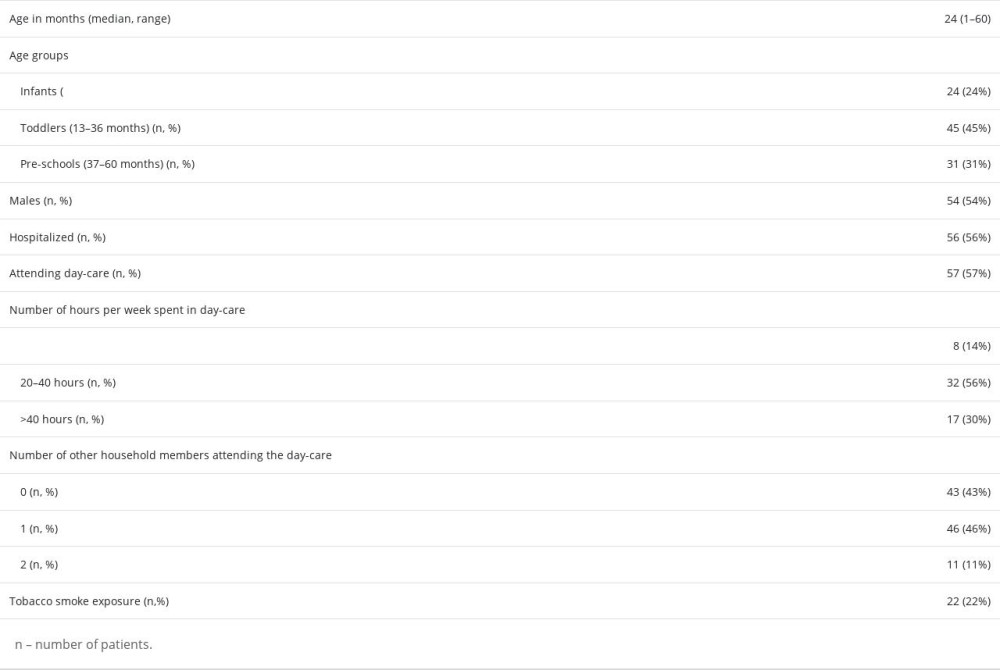 Table 1. Demographic data of children under age 5 with acute respiratory tract infections.
Table 1. Demographic data of children under age 5 with acute respiratory tract infections.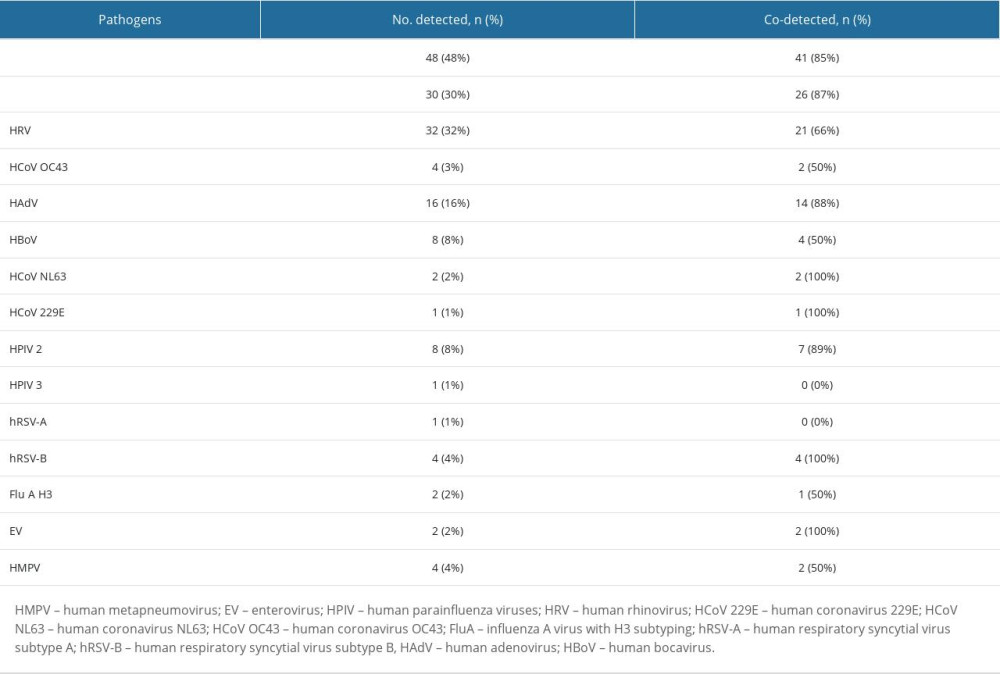 Table 2. Distribution of respiratory pathogens in children under age 5 with acute respiratory tract infections.
Table 2. Distribution of respiratory pathogens in children under age 5 with acute respiratory tract infections.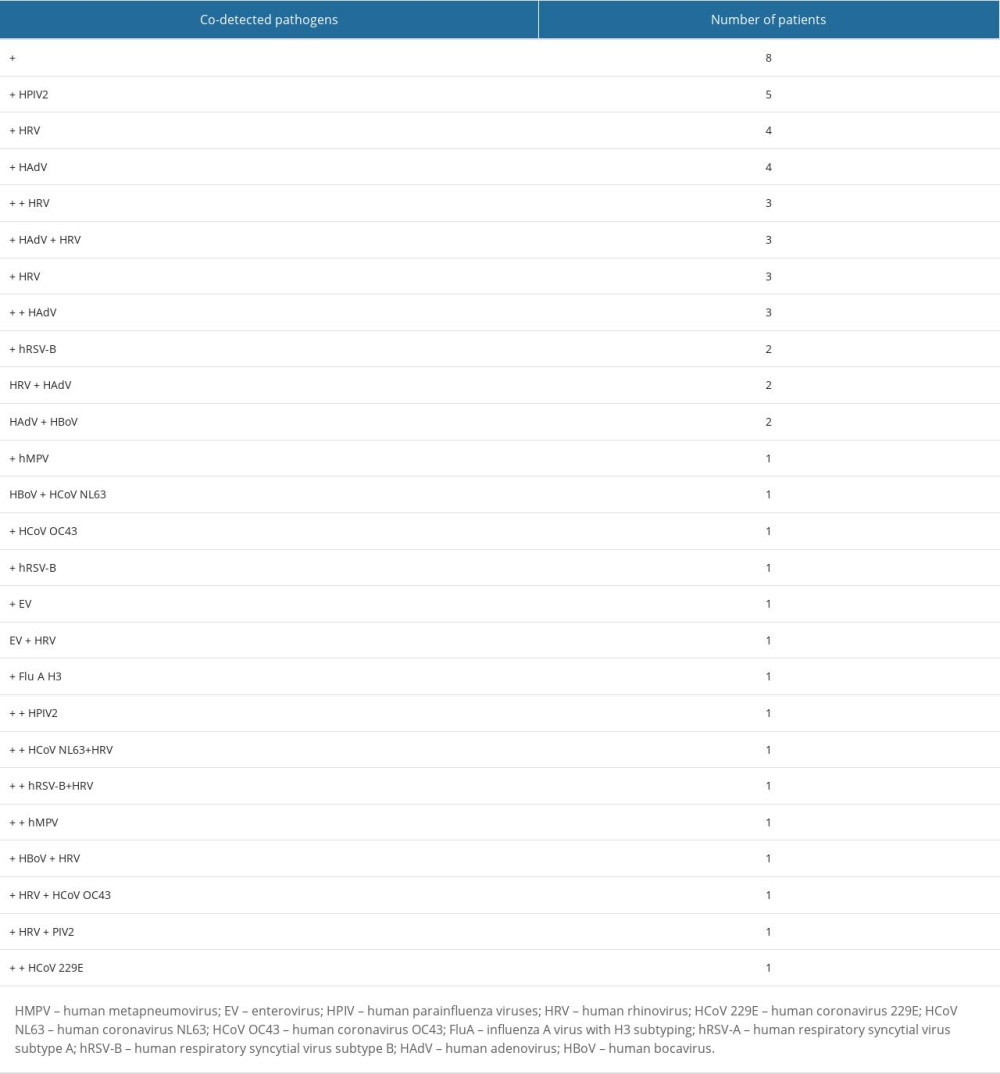 Table 3. Multiple detections in children with acute respiratory tract infections.
Table 3. Multiple detections in children with acute respiratory tract infections. Table 1. Demographic data of children under age 5 with acute respiratory tract infections.
Table 1. Demographic data of children under age 5 with acute respiratory tract infections. Table 2. Distribution of respiratory pathogens in children under age 5 with acute respiratory tract infections.
Table 2. Distribution of respiratory pathogens in children under age 5 with acute respiratory tract infections. Table 3. Multiple detections in children with acute respiratory tract infections.
Table 3. Multiple detections in children with acute respiratory tract infections. In Press
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
08 Mar 2024 : Laboratory Research
Evaluation of Retentive Strength of 50 Endodontically-Treated Single-Rooted Mandibular Second Premolars Res...Med Sci Monit In Press; DOI: 10.12659/MSM.944110
11 Mar 2024 : Clinical Research
Comparison of Effects of Sugammadex and Neostigmine on Postoperative Neuromuscular Blockade Recovery in Pat...Med Sci Monit In Press; DOI: 10.12659/MSM.942773
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








