05 October 2021: Clinical Research
miRNA Profiling of Hungarian Regressive Wilms’ Tumor Formalin-Fixed Paraffin-Embedded (FFPE) Samples by Quantitative Real-Time Polymerase Chain Reaction (RT-PCR)
Gergely Buglyó1ABCDEF*, Zsófia Magyar2ABCD, Éva Romicsné GörbeDOI: 10.12659/MSM.932731
Med Sci Monit 2021; 27:e932731
Abstract
BACKGROUND: Wilms’ tumor is a common renal malignancy of early childhood with a generally favorable prognosis depending upon histological subtype. It is becoming increasingly clear that differences in miRNA (microRNA) expression signature represent important clues helping us predict a tumor’s response to chemotherapy. In our study, we aimed to reveal miRNAs deregulated in regressive Wilms’ tumors from FFPE (formalin-fixed, paraffin-embedded) samples, also showing whether such samples are reliable miRNA sources in Wilms’ tumor.
MATERIAL AND METHODS: Samples from 8 Hungarian patients (3 males, 5 females, aged 1 to 7 years) were analyzed by qRT-PCR (quantitative real-time PCR). A PCR array was used in a pilot experiment, and selected miRNAs (miR-128-3p, miR-184, miR-194-5p, miR-203a) were studied in the rest of the samples using individual primers.
RESULTS: miR-194-5p was underexpressed in all tumor samples. miR-184 and miR-203a were underexpressed in 7 cases, the exception being a case with a high ratio of necrotic blastemal tissue. Results obtained with miR-128-3p are difficult to interpret due to varying directions of expression changes.
CONCLUSIONS: We conclude that a downregulation of miR-184, miR-194-5p, and miR-203a expression is observed in both regressive and blastemal tumors, but larger-scale studies are needed to confirm whether the degree of their underexpression correlates with the number of blastemal elements in a sample. In most of our FFPE samples aged up to 9 years, RNA extraction provided miRNA with quantity and quality sufficient for qRT-PCR-based analysis, emphasizing the relevance of pathological archives as miRNA sources in future studies.
Keywords: MIRN203 microRNA, Human, MIRN194 microRNA, Human, MIRN184 microRNA, Human, Real-Time Polymerase Chain Reaction, Wilms Tumor, Child, Child, Preschool, Female, Gene Expression Profiling, Humans, Hungary, Infant, Kidney Neoplasms, Paraffin Embedding
Background
Wilms’ tumor (nephroblastoma) is known to account for the vast majority (>90%) of childhood kidney malignancies, with an incidence ranging from 4.9 to 16.4 per 1 million children [1]. Management protocol recommended by SIOP (International Society of Paediatric Oncology) consists of 4–6 weeks of preoperative chemotherapy followed by surgical treatment. Postoperative treatment depends on the histology found in the surgical sample [2]. If more than two-thirds of the tumor mass shows chemotherapy-induced changes, the tumor is classified as regressive regardless of its histology. Non-regressive cases are further classified on the basis of the predominant cell type: blastemal, stromal, or epithelial. Focal or diffuse anaplastic changes may also occur and are associated with poor prognosis [3].
In non-anaplastic tumors, the presence of surviving blastemal tissue after preoperative chemotherapy is an important prognostic factor. According to the SIOP classification, regressive and blastemal are the 2 most common types of Wilms’ tumor, with the former showing the best long-term survival and the latter the poorest [4].
miRNAs (microRNAs) are a class of short, noncoding endogenous RNA molecules known to affect gene expression at the transcriptional and post-transcriptional level [5]. Their deregulation has been implicated by many authors in Wilms’ tumor, particularly the loss of let-7a and miR-200 family miRNAs due to mutations in miRNA processing genes [6,7] or the activity of oncogenes such as
Differences between miRNA signatures of blastemal and regressive Wilms’ tumors have been reported in the literature and are thought to be related to chemoresponsiveness [10]. In the present study, our aim was to find relevant, and possibly novel, miRNA deregulations from FFPE (formalin-fixed, paraffin-embedded) samples of a regressive Wilms’ tumor patient by PCR array, and study a narrower, hand-picked subset of miRNAs in the rest of our samples using individual primers, thereby contributing to our understanding of molecular factors behind the pathogenesis and chemoresponsiveness, and providing evidence for or against the feasibility of using FFPE samples for miRNA extraction.
Material and Methods
PATIENTS:
Our study population consisted of 10 white Hungarian patients diagnosed with unilateral regressive Wilms’ tumor at an age of 1 to 7 years, with a median age of 3 years (Table 1). Chemo- and surgical therapy were administered according to the SIOP WT 2001 protocol. MRI was used to assess tumor regression after completing preoperative chemotherapy, and the diagnosis of regressive Wilms’ tumor (and inclusion in the present study) was decided at this point. Patients 2 and 4 had triphasic tumors, meaning that tumor tissue contained viable blastemal, stromal, and epithelial cells after preoperative chemotherapy. Patient 8 was an unusual histological case in which blastemal Wilms’ tumor elements were necrotic, and viable parts of the neoplasm were mostly stromal. Patient 9 seemed to have multiple metastatic lesions in the lungs at the time of diagnosis, which were removed, but viable tumor cells were not observed in surgical samples. The final diagnosis was pulmonary fibrosis and the patient achieved remission. Patient 6 had a relapse resulting in local, intrapulmonary, and pleural metastases 8 months after diagnosis, which were surgically removed. Unfortunately, he did not show any response to subsequent adjuvant chemo- and radiotherapy, and he eventually died. All other patients remained in remission during follow-up.
SAMPLE HANDLING:
Analyzed FFPE samples were from children aged 5 to 9 years (median: 6.5 years) (Table 1). They were all prepared following the UMBRELLA protocol: surgical samples were subjected to immediate 24-h fixation in formaldehyde, then dehydrated by increasing concentrations of ethanol and embedded in paraffin wax. We needed 2 FFPE samples per patient – a case and a control – taken from the same kidney. As tumor samples, we used sections showing sufficient presence of non-necrotic tumor cells (this was not possible in Patients 9 and 10, forcing us to use necrotic tissue). Sections from adjacent tissue samples confirmed to be tumor-free were used as controls.
RNA EXTRACTION AND REVERSE TRANSCRIPTION:
We followed laboratory protocols as published before by our group and others [11,12]. We extracted miRNA from all samples using a miRNeasy FFPE Kit (Cat No. 217504) by Qiagen (Hilden, Germany), noted for its good performance [13]. Reverse transcription to obtain cDNA was performed using miScript II RT Kits (by Qiagen, Cat No. 218160). DNA was stored at −20°C for up to a few days until a PCR experiment was performed. Obtained miRNA and cDNA (complementary DNA) concentrations were adequate in 8 cases, but extraction was unsuccessful in 2 (both produced overwhelmingly necrotic tumor samples with a minimal number of viable cells remaining) which were excluded from further research (Table 1).
PILOT EXPERIMENT WITH QRT-PCR ARRAYS:
A pilot experiment with a single set of samples was designed to identify miRNA deregulations interesting enough to be studied in the rest of the samples. We used 2 SYBR Green-based 96-well miScript miRNA PCR Arrays (by Qiagen, Cat No.: 331221 MIHS-112ZF) containing, in addition to endogenous and exogenous controls, primers for 80 mature miRNAs and 4 miRNA precursors known as pathogenic in prostate cancer and presumed to be relevant in other types of genitourinary cancer such as Wilms’ tumor. Tumor and control cDNA samples from Patient 1 were amplified using the Roche LightCycler 96 PCR system. PCR reactions were initiated by a 15-min activation step at 95°C, consisting of 3-step cycling (denaturation for 15 s at 94°C, annealing for 30 s at 55°C, and extension for 30 s at 70°C) continued for 40 cycles. Data were evaluated by Roche’s own software LightCycler® 96, and analyzed according to the ΔΔCt method [14], using a single endogenous control gene SNORD61 (small nucleolar RNA U61) to ensure an easy comparison between the pilot arrays and later experiments. The reliability of SNORD61 as an endogenous control is supported both by the literature on malignancies of various origin [15,16] and by our own earlier data on Wilms’ tumor [11]. FC (fold change) is defined as 2−ΔΔCt [14], and FC values are shown on a logarithmic scale (hence, log2FC equals the opposite of ΔΔCt).
PCR EXPERIMENTS WITH INDIVIDUAL PRIMERS:
Assessing results from the pilot experiment and comparing them to literature data and to our previous results with non-regressive tumors [11], we chose 4 miRNAs to study in the rest of the samples: miR-128-3p, miR-184, miR-194-5p, and miR-203a (for the rationale of miRNA selection, see Discussion). Next, we used miScript Primer Assays provided by Qiagen for the 4 chosen miRNAs and our endogenous control SNORD61 (Cat No. MS00008582, MS00003640, MS00006727, MS00003766, and MS00033705, respectively). We amplified target sequences from tumor and control cDNA samples of each patient by the Roche LightCycler 96 PCR instrument also used for the pilot arrays. All other experimental (including PCR) conditions and reagents were the same as well, with 1 exception: 96-well PCR plates allowed for technical triplicates, so we used median Ct values to calculate ΔΔCt and fold change. Statistical tests were not performed because of the small sample size. This is an important limitation of our study.
DATABASE SEARCH:
Finally, we used MIENTURNET, a tool to search online databases to reveal and visualize a network of potential or validated miRNA targets [17]. Our query included miR-184, miR-194-5p, and miR-203a, while as a filter, we picked miRTarBase, a database containing experimentally validated targets only, with methods divided into 2 major categories: “strong” (eg, luciferase assays and Western blotting) and “weak” (eg, CLIP [cross-linking immunoprecipitation]).
COMPLIANCE WITH ETHICAL STANDARDS:
All procedures involving human participants were in accordance with the 1964 Helsinki Declaration and its later amendments and approved by an institutional ethics committee (approval statement No. SE RKEB 27/2020). Informed consent was obtained from the parents of all participants in the study.
Results
PILOT EXPERIMENT:
In Table 2, we demonstrate Ct values and fold changes for all precursor and mature miRNAs found on the PCR array we used in the pilot experiment. Results were adequate overall, but with an unusual overexpression of miR-133a, which we considered an artifact (see Discussion). miRNA precursor let-7a-5p was underexpressed with a fold change of −0.84 on the logarithmic scale. miRNAs of the miR-200 family also showed the expected degree of downregulation. Out of 80 mature miRNAs studied, 48 were substantially underexpressed (log2FC lower than −0.5) and 13 were overexpressed (log2FC higher than 0.5), while 19 were slightly or not changed (log2FC between −0.5 and 0.5).
EXPERIMENTS WITH INDIVIDUAL PRIMERS:
Expression data on other patients (IDs: 2 to 8) showed low variance among triplicates. Fold changes of miRNAs that were studied in all 8 patients – miR-128-3p, miR-184, miR-194-5p and miR-203a – are summarized in Table 3. miR-128-3p showed variable changes across samples (elevation in 4 cases, decrease in 3, minimal change in 1 case). Its mean fold change was close to 0. Levels of miR-184 were reduced in 7 cases and elevated in Patient 8. miR-194-5p was downregulated in all 8 cases. miR-203a was shown to be underexpressed in Patients 1 to 7 and overexpressed in Patient 8. Overall, the direction of some expression changes seems to have been reversed in Patient 8 (see Discussion).
NETWORK ANALYSIS:
We visualized targets of miR-184, miR-194-5p, and miR-203a, as obtained using the tool MIENTURNET (Figure 1). BMI1 (B cell-specific Moloney murine leukemia virus integration site 1) was shown to be affected by both miR-194-5p and miR-203a, while AKT2 (RAC-beta serine/threonine-protein kinase) was revealed as a common target of miR-184 and miR-203a.
Discussion
miRNAs, due to their short length, are thought to be stable enough to resist degradation and chemical modifications occurring in nucleic acids, especially RNA, as a result of formalin fixation. The quality of extracted miRNA was reported to be stable and unaffected by the preservation protocol used, the only exception being a 12-h delay to fixation, which resulted in a miRNA species-dependent variability in comparison to snap-frozen controls, possibly due to an increased breakdown of longer RNAs to products in the size range of miRNA [18]. Despite the wide availability of FFPE samples in pathological archives and a number of reports suggesting no difference between miRNA profiles obtained from snap-frozen tissue and FFPE samples from children aged 7 to 10 years with various types of cancer [13,19,20], miRNA-profiling from such samples has still not become a mainstream procedure.
Limitations of the present study include a small sample size preventing us from performing statistical analysis, and human bias in the selection of miRNAs for individual PCR experiments. Keeping these factors in mind, we interpret our findings below.
Our results with the pilot array were generally consistent with reports in the literature using frozen blood or tumor tissue as a miRNA source, with a few interesting findings discussed below. We considered the overexpression of miR-133a an artifact and did not include it in the Discussion, not only because of its magnitude, but also because miR-133a is known as a tumor suppressor in many cancer types [21,22], and has been reported to be unchanged [23] or underexpressed [11,24] in Wilms’ tumor.
In an earlier paper, we reported for the first time a miRNA expression analysis with RNA extracted from FFPE samples in Wilms’ tumor, focusing on the deregulation of miR-184, miR-194-5p, miR-203a, and miR-34c-5p [11]. Comparing these results obtained from blastemal tumors to our current pilot array data from regressive ones (Table 2), we decided to study the expression of a few miRNAs that showed interesting deregulations in the pilot experiment and were also featured in our previous paper – miR-184, miR-194-5p, and miR-203a – in all remaining patients, in order to reveal any differences between the pattern of their underexpression in blastemal and regressive tumors. Expression analysis of miR-34c-5p did not produce relevant findings in blastemal tumors, so we picked miR-128, which showed a promising elevation in our pilot array not yet reported in the literature, as the 4th miRNA to be studied. Other notable candidates included miR-205, which generally seems to play a tumor-suppressor role and is lost in some types of cancer [25,26], but the overexpression we observed had been reported before in Wilms’ tumor [23,24], and miR-29b, which was underexpressed in blastemal and regressive Wilms’ tumors, with some authors in support of this [27] and some reporting the opposite [23,24].
miR-128 is known to affect tumorigenesis in a number of cancer types [28] and has been suggested to play various regulatory roles in the central nervous system [29,30], but no reports are available linking it to the pathogenesis or prognosis of Wilms’ tumor. The overexpression we initially observed in Patient 1 was not reproducible across all samples, with some showing an elevation and others a decrease (Table 3).
miR-184, a tumor suppressor of various types of malignancy [31,32], was shown to inhibit autophagy and promote aging in rat glomerular mesangial cells [33]. As shown in Figure 1, its targets include GAS1 (Growth arrest-specific protein 1) [34] and AKT2, through which it suppresses tumorigenesis [35]. However, little is known about its possible function in the human kidneys, let alone in Wilms’ tumor. Earlier papers found it to be unchanged [23], underexpressed [27], or unaffected in anaplastic tumors, but downregulated in non-anaplastic tumors [36], while our own research revealed a pronounced downregulation in blastemal Wilms’ tumors [11]. We observed a similarly striking underexpression in our current set of regressive samples apart from Patient 8 (showing an unusual histology with very little persisting blastemal tissue and a slight elevation of miR-184) (Table 3).
A loss of miR-194-5p has long been known to be characteristic of Wilms’ tumors [23], but its role in the pathogenesis was shrouded in mystery until recently. Liu et al reported that miR-194-5p acts on the target gene
miR-203a is another important tumor suppressor with many targets, some shared with miR-184 and miR-194-5p (Figure 1). Earlier, we were the first group to report the loss of miR-203a in Wilms’ tumor [11]. Since then, other groups also noted its underexpression [36]. One study found it to be among the top 15 downregulated miRNAs (out of 156 deregulated miRNAs) in the disease [40], while another report claimed to have identified JAG1 (Jagged Canonical Notch Ligand 1) mRNA as its target, and suggested that a loss of miR-203a enhances a tumors’ migratory and invasive abilities through the upregulation of
In our set of regressive Wilms’ tumor samples, miR-203a underexpression was detectable in all cases except in Patient 8, who showed a nearly complete regression of blastemal tissue (Table 3). Overall expression of miR-203a, and to an extent that of miR-184 and miR-194-5p, appeared to be less reduced in regressive tumors compared to blastemal ones (Figure 2). It is also interesting to note the generally lower levels of miR-203a in samples containing more blastema, especially in the one that did not respond at all to chemotherapy [11]. Such an association, if confirmed by a larger-scale study in which statistical tests are applicable, may have prognostic and therapeutic implications.
Conclusions
Eighty-eight percent of all our Wilms’ tumor FFPE samples, including regressive specimens, which were the object of our current study and blastemal ones studied earlier, proved to be adequate for miRNA extraction (100% of samples containing visible non-necrotic tumor elements). We encourage other research groups to utilize the FFPE archives as miRNA sources in their projects involving Wilms’ tumor and other diseases. Despite the limitations, our results suggest that the degree of miR-203a underexpression is correlated with persisting blastemal fractions and prognosis of Wilms’ tumors. We plan to perform a larger-scale study to confirm this and reveal the role of other noncoding RNAs in the background of chemoresponsiveness.
Figures
![Network analysis showing targets of miR-184, miR-194-5p, and miR-203a from miRTarBase. A small network was constructed including targets validated by “strong” methods only, such as luciferase assays and western blots (A), while a larger network represents targets validated by either “strong” or “weak” (eg, CLIP) methods (B). Threshold set for the minimum number of miRNA-target interactions: 2. Created using the online tool MIENTURNET (version: N/A) [17].](https://jours.isi-science.com/imageXml.php?i=medscimonit-27-e932731-g001.jpg&idArt=932731&w=1000) Figure 1. Network analysis showing targets of miR-184, miR-194-5p, and miR-203a from miRTarBase. A small network was constructed including targets validated by “strong” methods only, such as luciferase assays and western blots (A), while a larger network represents targets validated by either “strong” or “weak” (eg, CLIP) methods (B). Threshold set for the minimum number of miRNA-target interactions: 2. Created using the online tool MIENTURNET (version: N/A) [17].
Figure 1. Network analysis showing targets of miR-184, miR-194-5p, and miR-203a from miRTarBase. A small network was constructed including targets validated by “strong” methods only, such as luciferase assays and western blots (A), while a larger network represents targets validated by either “strong” or “weak” (eg, CLIP) methods (B). Threshold set for the minimum number of miRNA-target interactions: 2. Created using the online tool MIENTURNET (version: N/A) [17]. ![A comparison of mean logarithmic fold changes (log2FC) in the expression of miR-184, miR-194-5p, and miR-203a between regressive Wilms’ tumor samples (light blue) and blastemal ones (deep blue, published earlier [11]). Created using Microsoft Excel (version: 2106).](https://jours.isi-science.com/imageXml.php?i=medscimonit-27-e932731-g002.jpg&idArt=932731&w=1000) Figure 2. A comparison of mean logarithmic fold changes (log2FC) in the expression of miR-184, miR-194-5p, and miR-203a between regressive Wilms’ tumor samples (light blue) and blastemal ones (deep blue, published earlier [11]). Created using Microsoft Excel (version: 2106).
Figure 2. A comparison of mean logarithmic fold changes (log2FC) in the expression of miR-184, miR-194-5p, and miR-203a between regressive Wilms’ tumor samples (light blue) and blastemal ones (deep blue, published earlier [11]). Created using Microsoft Excel (version: 2106). Tables
Table 1. Data on enrolled patients and FFPE samples with obtained miRNA concentrations.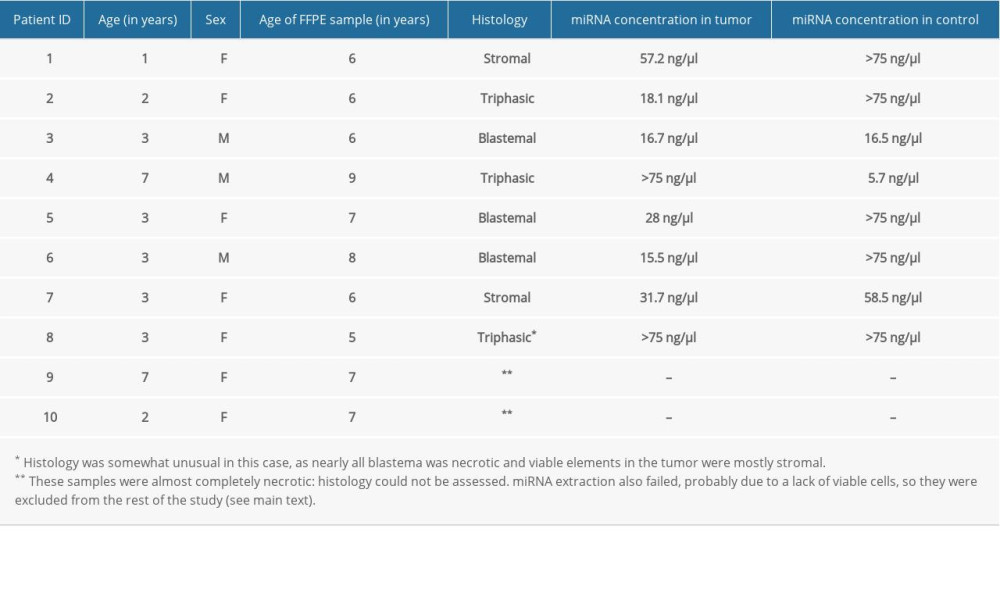 Table 2. Ct values and fold changes obtained in the pilot experiment by PCR array from tumor and control samples of Patient 1 using SNORD61 as endogenous control. Log2FC is the logarithm of fold change, defined as the opposite of ΔΔCt.
Table 2. Ct values and fold changes obtained in the pilot experiment by PCR array from tumor and control samples of Patient 1 using SNORD61 as endogenous control. Log2FC is the logarithm of fold change, defined as the opposite of ΔΔCt.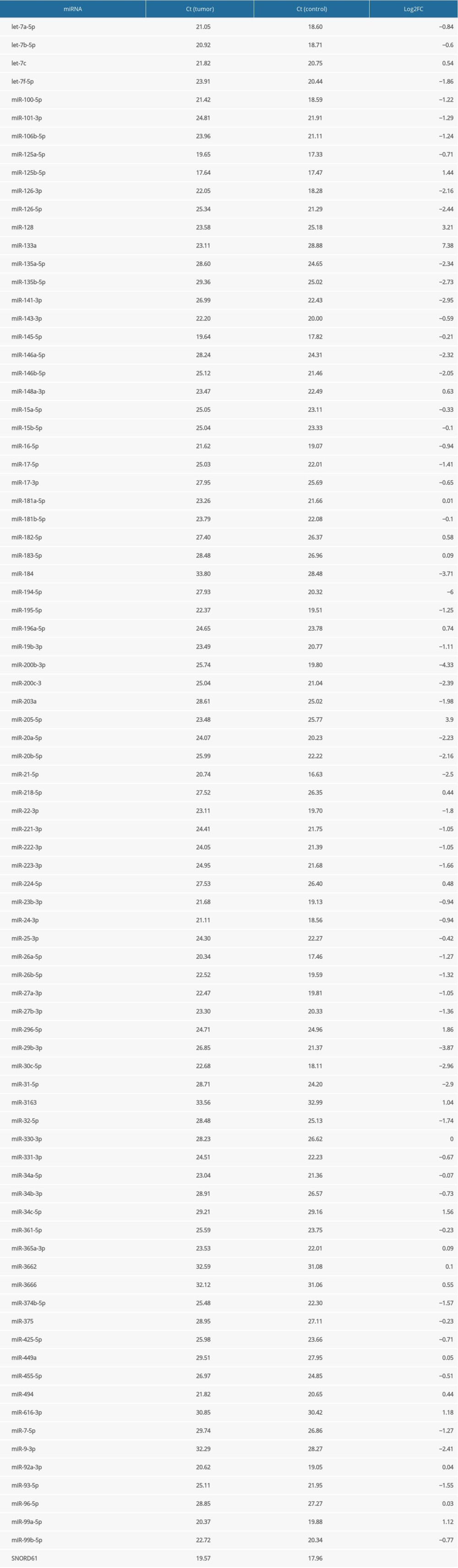 Table 3. Logarithmic fold changes (log2FC) of miR-128-3p, miR-184, miR-194-5p, and miR-203a in Patient 1 (determined by a PCR array), Patients 2 to 8 (determined by individual primers using the median value of 3 technical triplicates), and mean.
Table 3. Logarithmic fold changes (log2FC) of miR-128-3p, miR-184, miR-194-5p, and miR-203a in Patient 1 (determined by a PCR array), Patients 2 to 8 (determined by individual primers using the median value of 3 technical triplicates), and mean.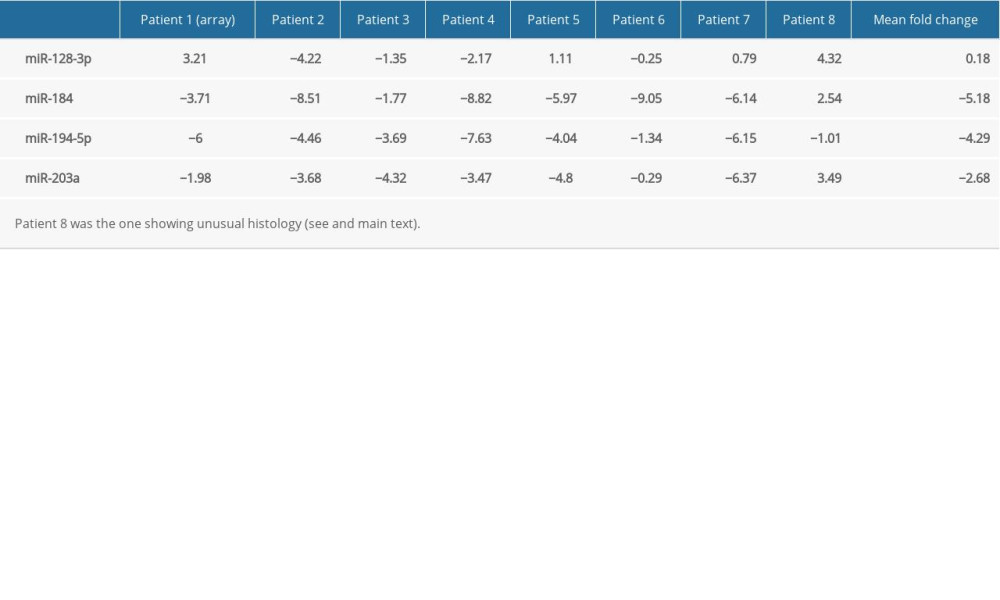
References
1. Cunningham ME, Klug TD, Nuchtern JG, Global disparities in Wilms tumor: J Surg Res, 2020; 247; 34-51
2. Bhatnagar S, Management of Wilms’ tumor: NWTS vs SIOP: J Indian Assoc Pediatr Surg, 2009; 14(1); 6-14
3. Phelps HM, Kaviany S, Borinstein SC, Lovvorn HN, Biological drivers of Wilms tumor prognosis and treatment: Children, 2018; 5(11); 145
4. Szychot E, Apps J, Pritchard-Jones K, Wilms’ tumor: Biology, diagnosis and treatment: Transl Pediatr, 2014; 3(1); 12-24
5. Pu M, Chen J, Tao Z, Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression: Cell Mol Life Sci, 2019; 76(3); 441-51
6. Gadd S, Huff V, Walz AL, Ooms AHAG, A Children’s Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor: Nat Genet, 2017; 49(10); 1487-94
7. Wegert J, Ishaque N, Vardapour R, Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors: Cancer Cell, 2015; 27(2); 298-311
8. Szemes M, Melegh Z, Bellamy J, Transcriptomic analyses of MYCN-regulated genes in anaplastic Wilms’ tumour cell lines reveals oncogenic pathways and potential therapeutic vulnerabilities: Cancers, 2021; 13(4); 656
9. Pode-Shakked N, Pleniceanu O, Gershon R, Dissecting stages of human kidney development and tumorigenesis with surface markers affords simple prospective purification of nephron stem cells: Sci Rep, 2016; 6; 23562
10. Watson JA, Bryan K, Williams R, miRNA profiles as a predictor of chemoresponsiveness in Wilms’ tumor blastema: PLoS One, 2013; 8(1); e53417
11. Buglyó G, Magyar Z, Romicsné Görbe É, Bánusz R, Quantitative RT-PCR-based miRNA profiling of blastemal Wilms’ tumors from formalin-fixed paraffin-embedded samples: J Biotechnol, 2019; 298; 11-15
12. Li Q, Li M, Zheng K, Detection of microRNA expression levels based on microarray analysis for classification of idiopathic pulmonary fibrosis: Exp Ther Med, 2020; 20(4); 3096-103
13. Howe K, Extraction of miRNAs from formalin-fixed paraffin-embedded (FFPE) tissues: Methods Mol Biol, 2017; 1509; 17-24
14. Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method: Methods, 2001; 25(4); 402-8
15. Sperveslage J, Hoffmeister M, Henopp T, Establishment of robust controls for the normalization of miRNA expression in neuroendocrine tumors of the ileum and pancreas: Endocrine, 2014; 46(2); 226-30
16. Zehentmayr F, Hauser-Kronberger C, Zellinger B, Hsa-miR-375 is a predictor of local control in early-stage breast cancer: Clin Epigenetics, 2016; 8; 28
17. Licursi V, Conte F, Fiscon G, Paci P, MIENTURNET: An interactive web tool for microRNA-target enrichment and network-based analysis: BMC Bioinformatics, 2019; 20(1); 545
18. Jones W, Greytak S, Odeh H, Deleterious effects of formalin-fixation and delays to fixation on RNA and miRNA-Seq profiles: Sci Rep, 2019; 9(1); 6980
19. Dijkstra JR, Mekenkamp LJM, Teerenstra S, MicroRNA expression in formalin-fixed paraffin embedded tissue using real time quantitative PCR: the strengths and pitfalls: J Cell Mol Med, 2012; 16(4); 683-90
20. Liu A, Xu X, MicroRNA isolation from formalin-fixed, paraffin-embedded tissues: Methods Mol Biol, 2011; 724; 259-67
21. Wang L-K, Hsiao T-H, Hong T-M, MicroRNA-133a suppresses multiple oncogenic membrane receptors and cell invasion in non-small cell lung carcinoma: PLoS One, 2014; 9(5); e96765
22. Chiyomaru T, Enokida H, Tatarano S, miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer: Br J Cancer, 2010; 102(5); 883-91
23. Liu M, Roth A, Yu M, The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis: Genes Dev, 2013; 27(23); 2543-48
24. Schmitt J, Backes C, Nourkami-Tutdibi N, Treatment-independent miRNA signature in blood of Wilms tumor patients: BMC Genomics, 2012; 13; 379
25. Hou S-X, Ding B-J, Li H-Z, Identification of microRNA-205 as a potential prognostic indicator for human glioma: J Clin Neurosci, 2013; 20(7); 933-37
26. Liu H, Lei C, He Q, Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer: Mol Cancer, 2018; 17(1); 64
27. Ludwig N, Werner TV, Backes C, Combining miRNA and mRNA expression profiles in Wilms tumor subtypes: Int J Mol Sci, 2016; 17(4); 475
28. Yao Y, Xu Q, Yan L, miRNA-128 and miRNA-142 regulate tumorigenesis and EMT in oral squamous cell carcinoma through HOXA10: Cancer Manag Res, 2020; 12; 9987-97
29. Zhang W, Kim PJ, Chen Z, MiRNA-128 regulates the proliferation and neurogenesis of neural precursors by targeting PCM1 in the developing cortex: Elife, 2016; 5; e11324
30. Zhou L, Yang L, Li Y-J, MicroRNA-128 protects dopamine neurons from apoptosis and upregulates the expression of excitatory amino acid transporter 4 in Parkinson’s disease by binding to AXIN1: Cell Physiol Biochem, 2018; 51(5); 2275-89
31. Zhen Y, Liu Z, Yang H, Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma: Cell Death Dis, 2013; 4; e872
32. Zhou R, Zhou X, Yin Z, Tumor invasion and metastasis regulated by microRNA-184 and microRNA-574-5p in small-cell lung cancer: Oncotarget, 2015; 6(42); 44609-22
33. Liu X, Fu B, Chen D, miR-184 and miR-150 promote renal glomerular mesangial cell aging by targeting Rab1a and Rab31: Exp Cell Res, 2015; 336(2); 192-203
34. Li W, Wang P, Zhang Z, MiR-184 regulates proliferation in nucleus pulposus cells by targeting GAS1: World Neurosurg, 2017; 97; 710-15.e1
35. Tu R, Chen Z, Bao Q, Liu H, Qing G, Crosstalk between oncogenic MYC and noncoding RNAs in cancer: Semin Cancer Biol, 2020 [Online ahead of print]
36. Pérez-Linares FJ, Pérezpeña-Diazconti M, García-Quintana J, MicroRNA profiling in Wilms tumor: Identification of potential biomarkers: Front Pediatr, 2020; 8; 337
37. Liu H, Ren S-Y, Qu Y, MiR-194-5p inhibited metastasis and EMT of nephroblastoma cells through targeting Crk: Kaohsiung J Med Sci, 2020; 36(4); 265-73
38. Zhou M, Xu Q, Huang D, Luo L, Regulation of gene transcription of B lymphoma Mo-MLV insertion region 1 homolog (review): Biomed Rep, 2021; 14(6); 52
39. Niu X, Zhu H-L, Liu Q, MiR-194-5p serves as a potential biomarker and regulates the proliferation and apoptosis of hippocampus neuron in children with temporal lobe epilepsy: J Chin Med Assoc, 2021; 84(5); 510-16
40. Zhang F, Zeng L, Cai Q, Comprehensive analysis of a long noncoding RNA-associated competing endogenous RNA network in Wilms tumor: Cancer Control, 2020; 27(2); 1073274820936991
41. Bao J-W, Li W-J, Guo J-H, MiRNA-203a-5p alleviates the malignant progression of Wilms’ tumor via targeting JAG1: Eur Rev Med Pharmacol Sci, 2020; 24(10); 5329-35
42. He J, Guo X, Sun L, Wang K, Yao H, Networks analysis of genes and microRNAs in human Wilms’ tumors: Oncol Lett, 2016; 12(5); 3579-85
43. Yang H, Wang L, Tang X, Bai W, miR-203a suppresses cell proliferation by targeting E2F transcription factor 3 in human gastric cancer: Oncol Lett, 2017; 14(6); 7687-90
44. Kort EJ, Farber L, Tretiakova M, The E2F3-Oncomir-1 axis is activated in Wilms’ tumor: Cancer Res, 2008; 68(11); 4034-38
45. An Q, Wang Y, An R, Association of E2F3 expression with clinicopathological features of Wilms’ tumors: J Pediatr Surg, 2013; 48(11); 2187-93
46. Bueno MJ, Gómez de Cedrón M, Laresgoiti U, Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling: Mol Cell Biol, 2010; 30(12); 2983-95
Figures
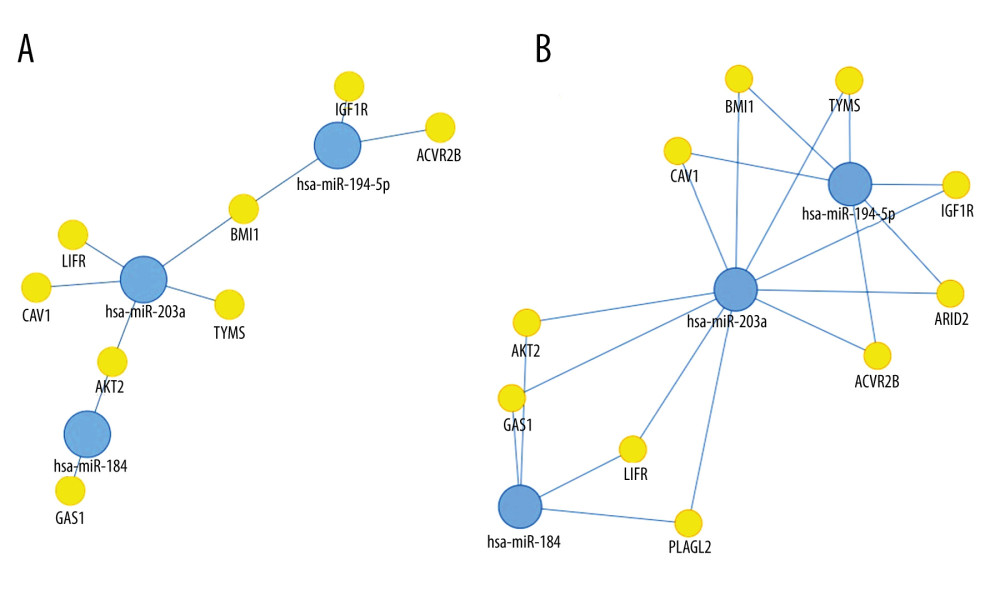 Figure 1. Network analysis showing targets of miR-184, miR-194-5p, and miR-203a from miRTarBase. A small network was constructed including targets validated by “strong” methods only, such as luciferase assays and western blots (A), while a larger network represents targets validated by either “strong” or “weak” (eg, CLIP) methods (B). Threshold set for the minimum number of miRNA-target interactions: 2. Created using the online tool MIENTURNET (version: N/A) [17].
Figure 1. Network analysis showing targets of miR-184, miR-194-5p, and miR-203a from miRTarBase. A small network was constructed including targets validated by “strong” methods only, such as luciferase assays and western blots (A), while a larger network represents targets validated by either “strong” or “weak” (eg, CLIP) methods (B). Threshold set for the minimum number of miRNA-target interactions: 2. Created using the online tool MIENTURNET (version: N/A) [17].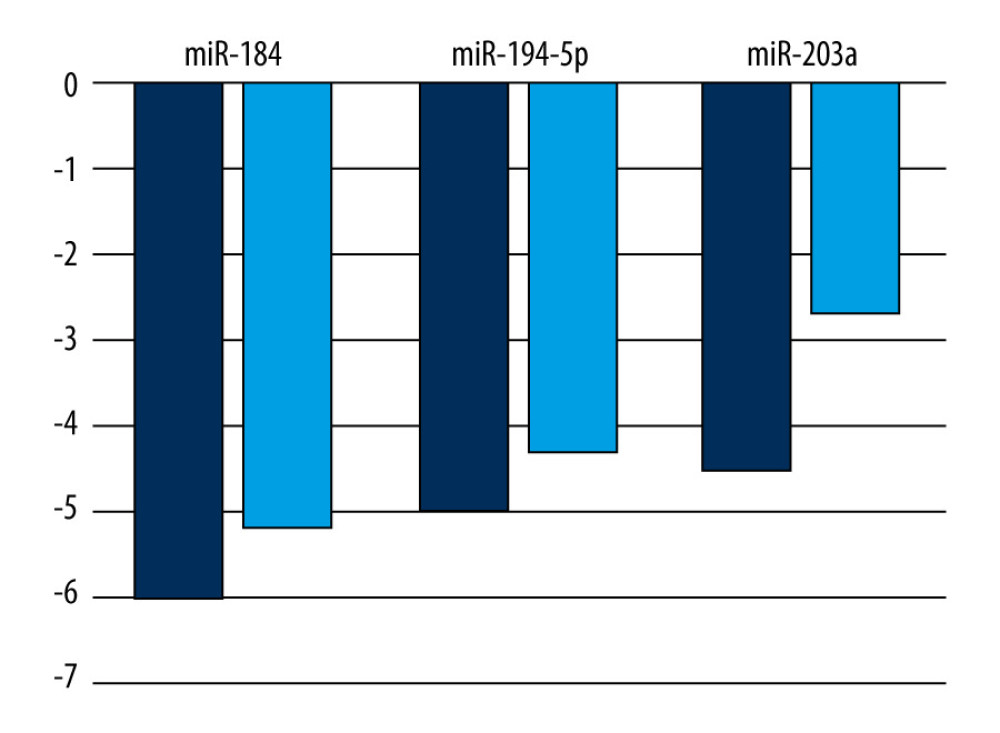 Figure 2. A comparison of mean logarithmic fold changes (log2FC) in the expression of miR-184, miR-194-5p, and miR-203a between regressive Wilms’ tumor samples (light blue) and blastemal ones (deep blue, published earlier [11]). Created using Microsoft Excel (version: 2106).
Figure 2. A comparison of mean logarithmic fold changes (log2FC) in the expression of miR-184, miR-194-5p, and miR-203a between regressive Wilms’ tumor samples (light blue) and blastemal ones (deep blue, published earlier [11]). Created using Microsoft Excel (version: 2106). Tables
 Table 1. Data on enrolled patients and FFPE samples with obtained miRNA concentrations.
Table 1. Data on enrolled patients and FFPE samples with obtained miRNA concentrations. Table 2. Ct values and fold changes obtained in the pilot experiment by PCR array from tumor and control samples of Patient 1 using SNORD61 as endogenous control. Log2FC is the logarithm of fold change, defined as the opposite of ΔΔCt.
Table 2. Ct values and fold changes obtained in the pilot experiment by PCR array from tumor and control samples of Patient 1 using SNORD61 as endogenous control. Log2FC is the logarithm of fold change, defined as the opposite of ΔΔCt. Table 3. Logarithmic fold changes (log2FC) of miR-128-3p, miR-184, miR-194-5p, and miR-203a in Patient 1 (determined by a PCR array), Patients 2 to 8 (determined by individual primers using the median value of 3 technical triplicates), and mean.
Table 3. Logarithmic fold changes (log2FC) of miR-128-3p, miR-184, miR-194-5p, and miR-203a in Patient 1 (determined by a PCR array), Patients 2 to 8 (determined by individual primers using the median value of 3 technical triplicates), and mean. Table 1. Data on enrolled patients and FFPE samples with obtained miRNA concentrations.
Table 1. Data on enrolled patients and FFPE samples with obtained miRNA concentrations. Table 2. Ct values and fold changes obtained in the pilot experiment by PCR array from tumor and control samples of Patient 1 using SNORD61 as endogenous control. Log2FC is the logarithm of fold change, defined as the opposite of ΔΔCt.
Table 2. Ct values and fold changes obtained in the pilot experiment by PCR array from tumor and control samples of Patient 1 using SNORD61 as endogenous control. Log2FC is the logarithm of fold change, defined as the opposite of ΔΔCt. Table 3. Logarithmic fold changes (log2FC) of miR-128-3p, miR-184, miR-194-5p, and miR-203a in Patient 1 (determined by a PCR array), Patients 2 to 8 (determined by individual primers using the median value of 3 technical triplicates), and mean.
Table 3. Logarithmic fold changes (log2FC) of miR-128-3p, miR-184, miR-194-5p, and miR-203a in Patient 1 (determined by a PCR array), Patients 2 to 8 (determined by individual primers using the median value of 3 technical triplicates), and mean. In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








