16 February 2021: Database Analysis
Overexpression of Apolipoprotein C1 (APOC1) in Clear Cell Renal Cell Carcinoma and Its Prognostic Significance
Huaying Xiao1CE, Yifang Xu2AE*DOI: 10.12659/MSM.929347
Med Sci Monit 2021; 27:e929347
Abstract
BACKGROUND: The aims of this study included 3 aspects: 1) assessing the expression of Apolipoprotein C1 (APOC1) in clear cell renal cell carcinoma (ccRCC) and normal groups; 2) evaluating the prognostic significance of APOC1 expression in the overall survival (OS) of ccRCC patients; and 3) exploring APOC1-related signaling pathways.
MATERIAL AND METHODS: The APOC1 expression value and clinical data of ccRCC patients were obtained from the cBioPortal database. We then evaluated the association of APOC1 expression with clinical characteristics of ccRCC patients. We also assessed the correlation between APOC1 expression and clinical outcome using Kaplan-Meier method. Our work then verified the independent prognostic factors of ccRCC by Cox regression analysis. Finally, the potential role of genes co-expressed with APOC1 was revealed via functional enrichment analysis.
RESULTS: Bioinformatic data revealed that APOC1 was expressed at higher levels in ccRCC tissue than in the normal group (all P<0.05). The high expression of APOC1 was associated with unfavorable prognosis of female patients (P<0.01), but not of male patients. APOC1 high expression also shortened the survival time of ccRCC patients age ≥60 years old (P<0.05). Cox regression analysis further indicated that APOC1 expression was an independent prognostic factor for OS of ccRCC patients. Additionally, we found that APOC1 expression was significantly associated with sex, grade, clinical stage, and T stage. Finally, enrichment analysis suggested that APOC1-associated pathways were involved in tumor growth and metastasis.
CONCLUSIONS: The current study indicated that APOC1 was highly expressed in ccRCC and was significantly associated with key clinical features. APOC1 appears to be an independent prognostic factor in patients with ccRCC. Importantly, APOC1 might be a potential therapeutic target for ccRCC via regulating pathways involved in cell growth and metastasis.
Keywords: Apolipoproteins C, Carcinoma, Renal Cell, Apolipoprotein C-I, Biomarkers, Tumor, Computational Biology, Databases, Genetic, Gene Expression Profiling, gene ontology, Gene Regulatory Networks, Kidney Neoplasms
Background
Renal cell carcinoma (RCC) is one of the most common malignancies of the urogenital system [1], and the incidence and mortality rates of RCC are increasing annually. Among RCCs, clear cell renal cell carcinoma (ccRCC) is the most common and highly malignant subtype of kidney cancer, accounting for about 70–85% of all cases [2]. Clinical surgery is currently recognized as the primary treatment for local ccRCC, but 20% of patients have postoperative recurrence despite local surgery [3]. Even worse, ccRCC is insensitive to radiotherapy and chemotherapy. Biological targeted therapy is becoming a research focus in cancer therapy. Therefore, it is of critical importance to assess the pathogenesis of ccRCC and find effective biomarkers to improve clinical outcomes of ccRCC patients.
Apolipoprotein C1 (APOC1) is the smallest apolipoprotein (6.6 KDa in size) of the apolipoprotein C family. The protein is mainly synthesized by the liver [4] and is highly expressed in this organ [5]; other organs, such as skin, lung, and intestines, also synthesize the protein. APOC1 is primarily found in chylomicrons, high-density lipoproteins (HDL), and very-low-density lipoproteins (VLDL) [6]. A previous study revealed that APOC1 is involved in numerous biological processes, including membrane remodeling, cholesterol catabolism, and dendritic reorganization [7]. In addition, APOC1 is associated with the development of type 2 diabetes, diabetic nephropathy, Alzheimer disease, and glomerulosclerosis [8–11]. Growing evidence also shows that APOC1 participates in the development and progression of cancers. It was revealed that APOC1 is highly expressed in gastric cancer (GC) [12], and high expression of APOC1 decreased the overall survival (OS) time of patients. APOC1 was also overexpressed in colorectal cancer (CC) tissue, which causes a poor prognosis [13]. This study further suggested that APOC1 is an independent risk factor for OS in CC. Moreover, APOC1 is associated with the development and prognosis of pancreatic cancer, prostate cancer [14], and lung cancer [15]. However, there have been few detailed investigations of the role of APOC1 in ccRCC.
In the present study, we first analyzed the APOC1 expression in ccRCC, and then determined the association between APOC1 expression and prognosis of ccRCC patients. Moreover, functional enrichment analyses were performed to investigate the role of APOC1 in ccRCC. We aimed to reveal the clinical significance of APOC1 in ccRCC, as well as to explore the possibility that APOC1 can be used as a predictive biomarker and molecular therapy target in ccRCC.
Material and Methods
APOC1 EXPRESSION IN VARIOUS CANCERS:
We obtained the transcription level of APOC1 in different cancers from the Oncomine database (
APOC1 EXPRESSION IN CCRCC:
The consistency of research can be verified by analysis of different databases. The differential expression of APOC1 in ccRCC and normal tissues was also assessed through the Gepia database (
ASSOCIATION OF APOC1 EXPRESSION WITH PROGNOSIS:
We initially evaluated the influence of APOC1 expression on the overall patient survival through Gepia by setting the expression median as the group cutoff. In order to obtain the detailed information, we conducted deeper survival analysis after considering several key clinical factors (sex, age, primary tumor laterality, histological grade, T stage, and clinical stage). To maintain the consistency in group cutoff with Gepia results, we defined the median of APOC1 expression level as the cutoff value, and assigned ccRCC patients into high- and low-expression groups. Kaplan-Meier (KM) analysis and log-rank test were subsequently used to evaluate the correlation between APOC1 expression (high vs low) and survival time of ccRCC patients. Finally, Cox regression analysis was performed to identify the independent prognosis factors of OS. It should be noted that some clinical data of patients were incomplete, which caused the inconsistency of sample sizes in survival analyses. The data of APOC1 expression value, age, and sex were available from 534 samples. However, among the 534 patients, 4 samples were lacking information of clinical stage and 8 samples lacked data on histological grade. Therefore, the sample sizes were inconsistent in survival analyses based on clinical stage and histological grade.
FUNCTIONAL ENRICHMENT ANALYSES:
For better understanding the role of APOC1 in ccRCC, the genes co-expressed with APOC1 were identified using the cBioPortal database. A total of 20 180 co-expressed genes were observed and 265 genes were finally selected for further enrichment analysis by setting q-value ≤0.05 and absolute value of Spearman’s r ≥0.5 as a threshold.
Bioinformatics tools, including FunRich and DAVID, were applied to perform enrichment analysis. Gene Ontology (GO) analysis was carried out with FunRich, enabling the annotation of cellular component (CC), biological process (BP), and molecular function (MF). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using DAVID (
STATISTICAL ANALYSIS:
SPSS 19.0 statistical software was used for clinical data analysis. APOC1 expressions in tumor and normal tissues were compared by Mann-Whitney U test. The independent-samples
Results
OVEREXPRESSION OF APOC1 IN CCRCC:
We first used the Oncomine database to visualize the expression of APOC1 in human cancers and normal specimens. Compared with normal tissues, the expression of APOC1 was mainly overexpressed in brain and central nervous system (CNS) cancer, breast cancer, lymphoma, kidney cancer, and gastric cancer (Figure 1A). Low expression of APOC1 was found in leukemia, lung cancer, and colorectal cancer. The results indicated that the expression of APOC1 was associated with cancer types.
The expression of APOC1 in kidney cancer was further explored. All published research indicated the high expression of APOC1 in kidney cancer. As ccRCC is the most common subtype of kidney cancer, 6 datasets reporting ccRCC were finally selected to perform meta-analysis in the Oncomine database. The results disclosed that APOC1 was significantly upregulated in all ccRCC tissues (Figure 1B). Details of the differences in APOC1 expression between tumor and normal tissues among 6 studies are presented with box plots (Figure 2A–2F). Research information of 6 datasets, including fold change, P value, and sample size, are summarized in Table 1.
ASSOCIATION OF APOC1 EXPRESSION WITH CLINICAL PROGNOSIS:
The APOC1 expressions in clinical tumor and normal tissues were further verified in the HPA database. The results indicated the overexpression of APOC1 in tumor tissues (Figure 3A), which was consistent with the results in Gepia (Figure 3B). To assess the significance of APOC1 expression on the prognosis of patients, KaplanMeier analysis was performed to estimate the correlation between APOC1 expression and overall survival. The survival analysis showed that high expression of APOC1 was associated with shorter survival time and poor prognosis of patients with kidney cancer (Figure 3C, P=0.024).
Due to the important association of sex and age with clinical outcome, these 2 factors were assessed in survival analysis. Our result suggested that the survival time was similar in male and female patients (Figure 4A), high expression of APOC1 was associated with poor prognosis of female patients (P=0.001), but APOC1 high expression was not associated with male survival (P=0.601). Survival time was associated by the age of patients (Figure 4B), high expression of APOC1 was associated with shorter survival time of ccRCC patients age ≥60 years (P=0.032), OS was not different in patients with age <60 years (P=0.542). As expected, the clinical prognosis of ccRCC patients was associated with clinical stage (Figure 4C, P<0.001) and histological grade of patients (Figure 4D, P<0.001).
The sensitivity and specificity of the APOC1 expression for OS and DFS predictions were assessed by ROC analysis. Our results indicated that APOC1 achieved a moderate predicting power for OS, with an AUC of 0.595 (Figure 5A, P<0.001). For DFS prediction, ROC analysis showed an AUC of 0.591 (Figure 5B, P=0.003). Thus, APOC1 may be used as a novel and moderate prognostic biomarker for predicting the OS and DFS of patients with ccRCC.
There were several clinical factors influencing the prognosis of patients. Univariate analysis indicated that sex was not associated with the survival of patients, and other parameters were all significantly associated with OS of ccRCC patients (all P<0.05). When integrating these factors into multivariate Cox regression analysis, APOC1 expression (HR=1.379, P=0.036), age (HR=1.802, P<0.001), clinical stage (HR=3.563, P<0.001), and histological grade (HR=1.818, P=0.001) remained as independent prognostic factors for OS in patients with ccRCC (Table 2).
ASSOCIATION OF APOC1 EXPRESSION WITH CLINICOPATHOLOGICAL FACTORS:
The above results show that the expression of APOC1 exerts a vital role on the prognosis of patients. Therefore, we further evaluated the factors affecting APOC1 expression. Figure 6A shows that expression of APOC1 was associated with sex, and higher expression was observed in male patients (P<0.001). Age (Figure 6C) and tumor sites (Figure 6B) did not influence APOC1 expression. For histological grade (Figure 6D), APOC1 expression gradually increased with high grade. Regarding T stage (Figure 6E), the lowest expression of APOC1 appeared in T1, which was significantly smaller than that in T2 stage (P<0.01) and T3 stage (P<0.001). When it came to clinical stage (Figure 6F), APOC1 expressions in stage III and IV were higher than that in stage I (all P<0.001). Taken together, these results indicated that the expression of APOC1 was associated with many key clinical features of ccRCC.
ENRICHMENT ANALYSIS OF GENES CO-EXPRESSED WITH APOC1:
To further elucidate the potential role of APOC1 in ccRCC, we first determined the genes that are co-expressed with APOC1. In our study, a total of 265 co-expressed genes with APOC1 were finally selected by setting q-value ≤0.05 and absolute value of Spearman’s r ≥0.5 as threshold. We then carried out functional enrichment analysis using FunRich software. It was found that these co-expressed genes were mainly enriched in dendritic cell, leukocytes, B cell, CD4, and ascites cancer cell (Figure 7), suggesting that these genes participate in immune response.
The 265 co-expressed genes were also used for GO enrichment analysis, and the top 10 enrichment results for molecular function (MF), biological process (BP), and cellular component (CC) were presented. MF analysis showed that these genes were mainly enriched in receptor activity, receptor signaling complex scaffold activity, and MHC class I and II receptor activities (Figure 8A). For BP, co-expressed genes were significantly enriched in immune response (Figure 8B). With regard to CC, co-expressed genes were significantly enriched in plasma membrane, exosomes, NADPH oxidase complex, lysosome, and extracellular region (Figure 8C).
We finally performed KEGG pathway analysis to investigate the relevant pathways of these genes co-expressed in ccRCC. We found that these genes significantly participated in 34 pathways (P<0.05), and the top 10 pathways are shown in Figure 8D. The most significant pathways were Staphylococcus aureus infection, osteoclast differentiation, lysosome, phagosome, tuberculosis, chemokine signaling pathway, fc gamma R-mediated phagocytosis, antigen processing and presentation, viral myocarditis, systematic lupus, and erythematosus. Combining P value and percent of genes, the top 5 pathways were enriched in osteoclast differentiation, lysosome, phagosome, tuberculosis, and chemokine signaling pathway.
Discussion
The present study revealed that the expression of APOC1 was associated with sex, age, histological grade, T stage, and clinical stage. High expression of APOC1 was observed in ccRCC patients and was associated with poor prognosis. Cox regression analysis suggested that APOC1 expression remained as an independent prognostic factor for OS of ccRCC patients. A recent study by Cui et al revealed that high expression of APOC1 was correlated with shorter overall survival (
A previous study speculated that APOC1 may act as a tumor promoter by regulating cholesterol metabolism, and APOC1 might play a key role in the tumorigenesis and progression of ccRCC [16]. A study by Li et al indicated that APOC1 acts as a novel pro-metastatic factor, which facilitated the activation of STAT3 and enhanced the metastasis of ccRCC cells [17]. These studies contributed to our understanding of the potential mechanism of APOC1 in ccRCC. Our study also explored the APOC1-related pathway involved in ccRCC by performing KEGG pathway analysis. We found that most of the significant pathways involved the process of proliferation and metastasis of ccRCC. Osteoclast differentiation, lysosomes, and phagosome were associated with tumor metastasis. Bone metastasis has become a common phenomenon in the late stage of cancers. In the process of bone metastasis, cancer cells promote osteoclast differentiation and activation, which in turn induces the seeding and proliferation of cancer cells in bone4[18]. Inhibition of bone metastasis is of vital importance in reducing the incidence of skeletal complications and improving patient quality of life [19]. Lysosomes are emerging as a central regulator of the autophagic process, and play a critical role in tumor growth [20]. Lysosome-mediated autophagy has been considered as an effective mechanism for cancer cell survival, and degradation of lysosomes is regarded as a new targeted cancer treatment [21]. Lysosome trafficking is a key event in the process of metastasis and plays a significant role in tumor invasion. In addition, apoptosis-mediated immune response is involved in the development of ccRCC. For example,
Besides proliferation and metastasis, there are several undesirable factors participating in the development of kidney disease. For instance, patients with systematic lupus and erythematosus appear to be at increased risk of cancer, particularly non-Hodgkin lymphoma and kidney cancer [24]. Tuberculosis has been microbiologically demonstrated to be an etiology of membranous nephropathy and interstitial nephritis [25], which greatly increases the risk of fatal kidney cancer (HR=6.16) [26]. These investigations indicate that tuberculosis is closely correlated with kidney disease.
To understand the function of APOC1 in ccRCC, we performed GO analysis, which indicated that molecular function focused on receptor activities, such as MHC class I and II receptor activities. Biological process was mainly involved in immune response, and cellular component involved plasma membranes, exosomes, and lysosomes. These genes co-expressed with APOC1 were mainly expressed in dendritic cells, B cells, and CD4 cells. Dendritic cells are the most important antigen-presenting cells, which can present antigens via both MHC-I and MHC-II [27], and play a primary role in antigen presentation to CD4 cells. Dendritic cells can determine cancer immune responses by regulating immune activation and tolerance [28], as well as leading to the development of autoantibody through B cells [29]. The present results are consistent with the literature.
Our study has certain limitations that should be considered. 1) Some clinical data on patient prognosis were incomplete. 2) The expression of APOC1 in tumor and normal tissues should be verified in clinical samples. 3) The underlying molecular mechanisms indicated by pathway analysis have not been determined yet. 4) All of the prognoses were based on mRNA level, and all of the data were from bioinformatics, which weakens the strength of our conclusions. Further experimental research is needed to support our findings. However, our results may be valuable to clinicians and researchers, demonstrating a new biomarker for kidney cancer, which may help develop novel early interventions in cancer treatment.
Conclusions
This study explored the role of APOC1 in ccRCC by using bioinformatics data mining. It was revealed that APOC1 was highly expressed in ccRCC tissue, and high expression of APOC1 was associated with poor prognosis of patients. APOC1 remained an independent prognostic factor for OS in ccRCC patients. Moreover, the co-expressed genes with APOC1 were significantly enriched in pathways relating to tumor growth and metastasis. Taken together, our results suggest APOC1 is a potential therapeutic target for ccRCC treatment in the future.
Figures
 Figure 1. Expression of APOC1 in human cancers. (A) Disease summary for APOC1. red: high expression, blue: low expression. The threshold was set as follows: P value: 0.05, fold change: 2, gene rank: 10%, (B) Comparison of APOC1 across 6 analyses regarding ccRCC via meta-analysis. All data were derived from the Oncomine database based on TCGA data.
Figure 1. Expression of APOC1 in human cancers. (A) Disease summary for APOC1. red: high expression, blue: low expression. The threshold was set as follows: P value: 0.05, fold change: 2, gene rank: 10%, (B) Comparison of APOC1 across 6 analyses regarding ccRCC via meta-analysis. All data were derived from the Oncomine database based on TCGA data. 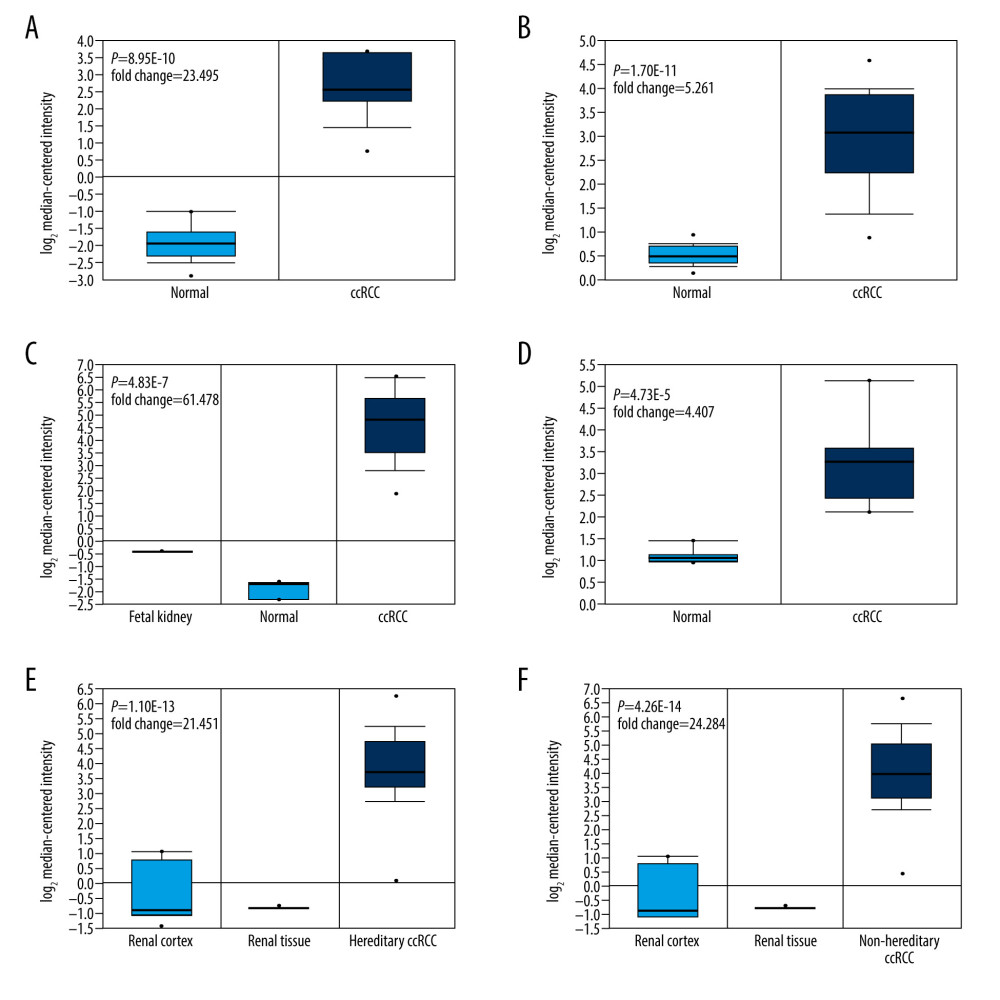 Figure 2. (A–F) The differential analyses of APOC1 expression in different datasets. All data were derived from the Oncomine database based on TCGA data.
Figure 2. (A–F) The differential analyses of APOC1 expression in different datasets. All data were derived from the Oncomine database based on TCGA data. 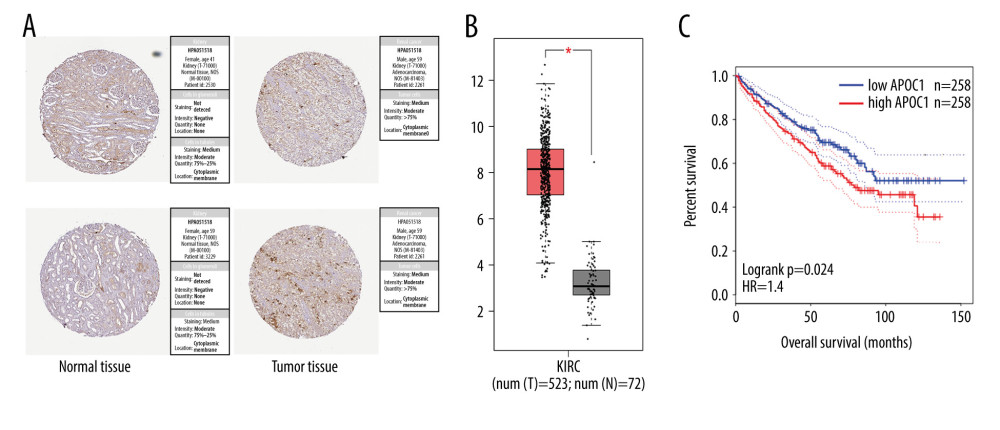 Figure 3. APOC1 expression and prognosis analyses in kidney renal clear cell carcinoma. (A) Representative immunohistochemistry images and detailed information on APOC1 in kidney cancer and normal tissues that were obtained from the HPA database; (B) Tumor/normal differential expression analysis in Gepia; (C) Patient survival analysis in Gepia.
Figure 3. APOC1 expression and prognosis analyses in kidney renal clear cell carcinoma. (A) Representative immunohistochemistry images and detailed information on APOC1 in kidney cancer and normal tissues that were obtained from the HPA database; (B) Tumor/normal differential expression analysis in Gepia; (C) Patient survival analysis in Gepia. 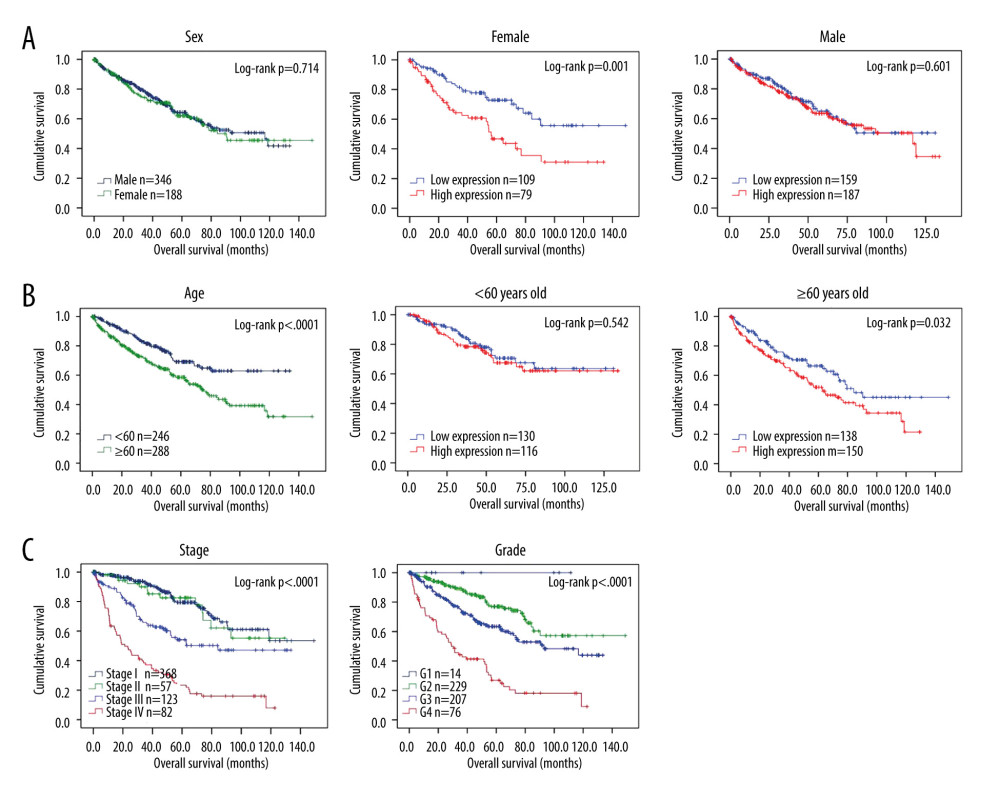 Figure 4. Survival analyses based on clinical characteristics of ccRCC patients. (A) Sex; (B) Age; (C) Clinical stage; (D) Histological grade.
Figure 4. Survival analyses based on clinical characteristics of ccRCC patients. (A) Sex; (B) Age; (C) Clinical stage; (D) Histological grade. 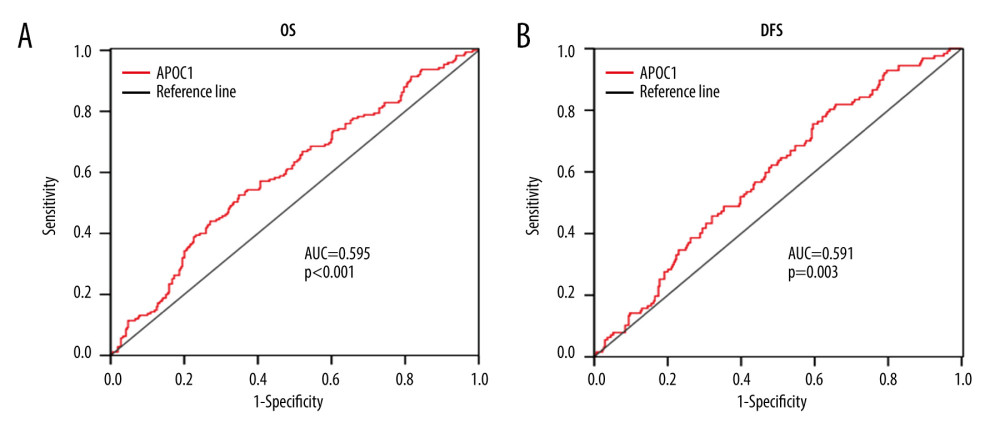 Figure 5. ROC analysis for predicting prognosis. (A) Overall survival (OS); (B) Disease-free survival (DFS).
Figure 5. ROC analysis for predicting prognosis. (A) Overall survival (OS); (B) Disease-free survival (DFS). 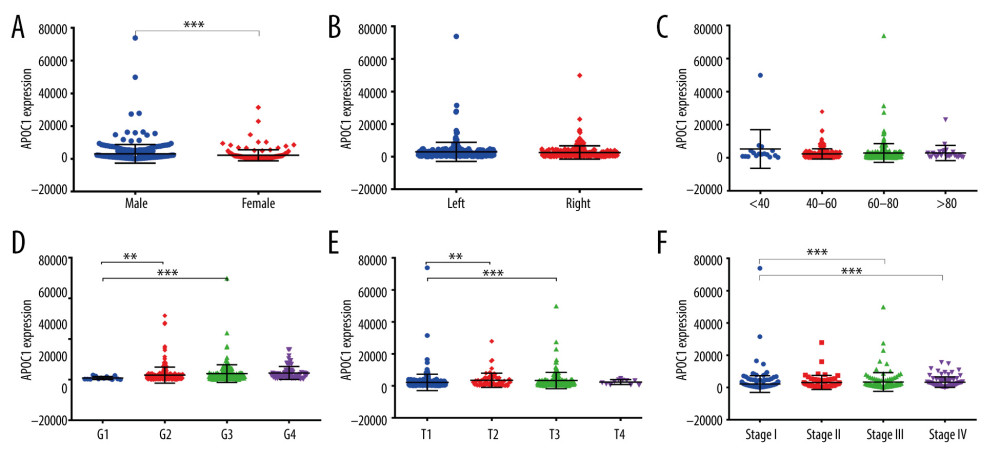 Figure 6. APOC1 expression analyses based on clinicopathological factors. (A) Sex; (B) Primary tumor laterality; (C) Age; (D) Histological grade; (E) T stage; (F) Clinical stage. Compared with normal group: ** P<0.01 and *** P<0.001.
Figure 6. APOC1 expression analyses based on clinicopathological factors. (A) Sex; (B) Primary tumor laterality; (C) Age; (D) Histological grade; (E) T stage; (F) Clinical stage. Compared with normal group: ** P<0.01 and *** P<0.001. 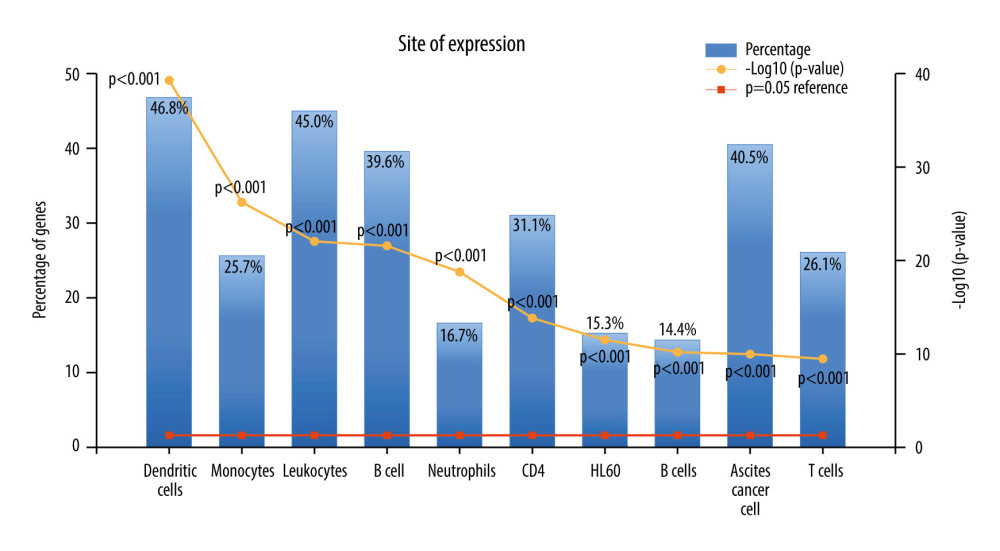 Figure 7. The expression of co-expressed genes with APOC1 in various cells.
Figure 7. The expression of co-expressed genes with APOC1 in various cells. 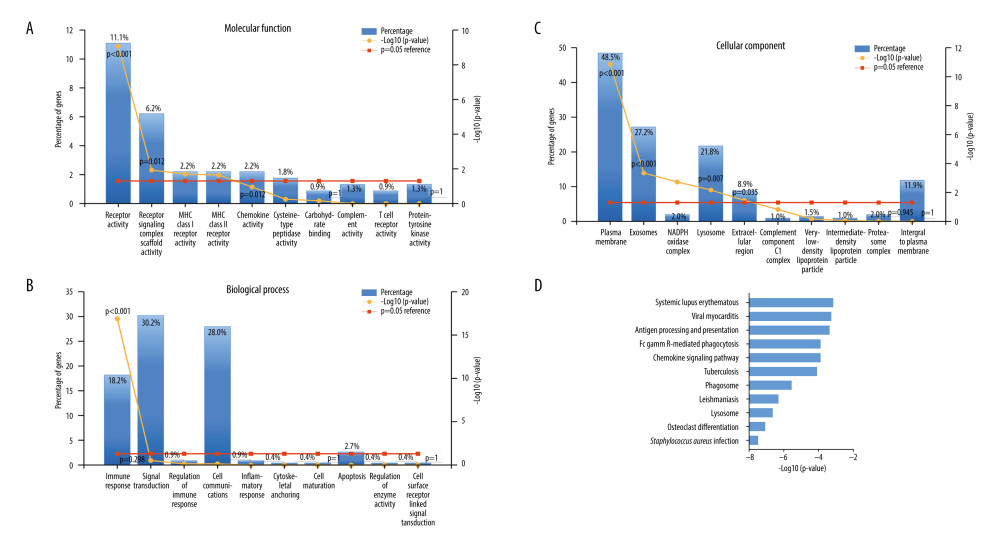 Figure 8. GO annotation and KEGG pathway analyses. (A) Molecular function (MF); (B) Biological process (BP); (C) Cellular component (CC); (D) Biological pathways.
Figure 8. GO annotation and KEGG pathway analyses. (A) Molecular function (MF); (B) Biological process (BP); (C) Cellular component (CC); (D) Biological pathways. References
1. Wang J, Xu Y, Zhu L, Cannabinoid receptor 2 as a novel target for promotion of renal cell carcinoma prognosis and progression: J Cancer Res Clin Oncol, 2018; 144; 39-52
2. Xu WH, Xu Y, Tian X, Anwaier A, Large-scale transcriptome profiles reveal robust 20-signatures metabolic prediction models and novel role of G6PC in clear cell renal cell carcinoma: J Cell Mol Med, 2020; 24(16); 9012-27
3. Pang LR, Huang MX, Li H, LINC00707 accelerates the proliferation, migration and invasion of clear cell renal cell carcinoma: Eur Rev Med Pharmacol Sci, 2020; 24; 6616-22
4. Jong MC, Hofker MH, Havekes LM, Role of ApoCs in lipoprotein metabolism: Functional differences between ApoC1, ApoC2, and ApoC3: Arterioscler Thromb Vasc Biol, 1999; 19; 472-84
5. Jong MC, Gijbels MJ, Dahlmans VE, Hyperlipidemia and cutaneous abnormalities in transgenic mice overexpressing human apolipoprotein C1: J Clin Invest, 1998; 101; 145-52
6. Brewer HB, Shulman R, Herbert P, The complete amino acid sequence of alanine apolipoprotein (apoC-3), and apolipoprotein from human plasma very low density lipoproteins: J Biol Chem, 1974; 249; 4975-84
7. Leduc V, Jasmin-Belanger S, Poirier J, APOE and cholesterol homeostasis in Alzheimer’s disease: Trends Mol Med, 2010; 16; 469-77
8. Mooyaart AL, Valk EJ, van Es LA, Genetic associations in diabetic nephropathy: A meta-analysis: Diabetologia, 2011; 54; 544-53
9. McKay GJ, Savage DA, Patterson CC, Association analysis of dyslipidemia-related genes in diabetic nephropathy: PLoS One, 2013; 8; e58472
10. Stagg RJ, Venook AP, Chase JL, Alternating hepatic intra-arterial floxuridine and fluorouracil: A less toxic regimen for treatment of liver metastases from colorectal cancer: J Natl Cancer Inst, 1991; 83; 423-28
11. Bus P, Pierneef L, Bor R, Apolipoprotein C-I plays a role in the pathogenesis of glomerulosclerosis: J Pathol, 2017; 241; 589-99
12. Yi J, Ren L, Wu J, Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for gastric cancer: Ann Transl Med, 2019; 7; 380
13. Ren H, Chen Z, Yang L, Apolipoprotein C1 (APOC1) promotes tumor progression via MAPK signaling pathways in colorectal cancer: Cancer Manag Res, 2019; 11; 4917-30
14. Su WP, Sun LN, Yang SL, Apolipoprotein C1 promotes prostate cancer cell proliferation in vitro: J Biochem Mol Toxicol, 2018; 32; e22158
15. Hughes BA, Fleming CR, Berkner S, Gaffron R, Patient compliance with a home parenteral nutrition program: JPEN J Parenter Enteral Nutr, 1980; 4; 12-14
16. Cui Y, Miao C, Hou C, Apolipoprotein C1 (APOC1):A novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma: Front Oncol, 2020; 10; 1436
17. Li Y, Wu L, Zeng L, ApoC1 promotes the metastasis of clear cell renal cell carcinoma via activation of STAT3: Oncogene, 2020; 39; 6203-17
18. Taverna S, Giusti I, D’Ascenzo S, Breast cancer derived extracellular vesicles in bone metastasis induction and their clinical implications as biomarkers: Int J Mol Sci, 2020; 21; 3573
19. Takahashi S, Kinuya S, Nonomura N, Japanese Expert Panel Meeting on the management of prostate cancer with bone metastases: Oncol Ther, 2018; 6; 157-71
20. He R, Shi X, Zhou M, Alantolactone induces apoptosis and improves chemosensitivity of pancreatic cancer cells by impairment of autophagy-lysosome pathway via targeting TFEB: Toxicol Appl Pharmacol, 2018; 356; 159-71
21. Purushothaman B, Lee J, Hong S, Song JM, Multifunctional TPP-PEG-biotin self-assembled nanoparticle drug delivery-based combination therapeutic approach for co-targeting of GRP78 and lysosome: J Nanobiotechnology, 2020; 18; 102
22. Nasser A, Azimi T, Ostadmohammadi S, Ostadmohammadi S: Microb Pathog, 2020; 148; 104431
23. Torchinsky MB, Garaude J, Blander JM, Infection and apoptosis as a combined inflammatory trigger: Curr Opin Immunol, 2010; 22; 55-62
24. Tallbacka KR, Pettersson T, Pukkala E, Increased incidence of cancer in systemic lupus erythematosus: A finnish cohort study with more than 25 years of follow-up: Scand J Rheumatol, 2018; 47; 461-64
25. Malhotra KP, Chandra A, Rao N, Tuberculosis as a microbiologically proven etiology of membranous nephropathy and interstitial nephritis: Saudi J Kidney Dis Transpl, 2019; 30; 1447-49
26. Su VY, Yen YF, Pan SW, Latent tuberculosis infection and the risk of subsequent cancer: Medicine (Baltimore), 2016; 95; e2352
27. Gholizadeh Z, Tavakkol-Afshari J, Nikpoor AR, Enhanced immune response induced by P5 HER2/neu-derived peptide-pulsed dendritic cells as a preventive cancer vaccine: J Cell Mol Med, 2018; 22; 558-67
28. Qian C, Yang LJ, Cui H, Recent advances in nanotechnology for dendritic cell-based immunotherapy: Front Pharmacol, 2020; 11; 960
29. Yu H, Liu Y, Han J, TLR7 regulates dendritic cell-dependent B-cell responses through BlyS in immune thrombocytopenic purpura: Eur J Haematol, 2011; 86; 67-74
Figures
 Figure 1. Expression of APOC1 in human cancers. (A) Disease summary for APOC1. red: high expression, blue: low expression. The threshold was set as follows: P value: 0.05, fold change: 2, gene rank: 10%, (B) Comparison of APOC1 across 6 analyses regarding ccRCC via meta-analysis. All data were derived from the Oncomine database based on TCGA data.
Figure 1. Expression of APOC1 in human cancers. (A) Disease summary for APOC1. red: high expression, blue: low expression. The threshold was set as follows: P value: 0.05, fold change: 2, gene rank: 10%, (B) Comparison of APOC1 across 6 analyses regarding ccRCC via meta-analysis. All data were derived from the Oncomine database based on TCGA data. Figure 2. (A–F) The differential analyses of APOC1 expression in different datasets. All data were derived from the Oncomine database based on TCGA data.
Figure 2. (A–F) The differential analyses of APOC1 expression in different datasets. All data were derived from the Oncomine database based on TCGA data. Figure 3. APOC1 expression and prognosis analyses in kidney renal clear cell carcinoma. (A) Representative immunohistochemistry images and detailed information on APOC1 in kidney cancer and normal tissues that were obtained from the HPA database; (B) Tumor/normal differential expression analysis in Gepia; (C) Patient survival analysis in Gepia.
Figure 3. APOC1 expression and prognosis analyses in kidney renal clear cell carcinoma. (A) Representative immunohistochemistry images and detailed information on APOC1 in kidney cancer and normal tissues that were obtained from the HPA database; (B) Tumor/normal differential expression analysis in Gepia; (C) Patient survival analysis in Gepia. Figure 4. Survival analyses based on clinical characteristics of ccRCC patients. (A) Sex; (B) Age; (C) Clinical stage; (D) Histological grade.
Figure 4. Survival analyses based on clinical characteristics of ccRCC patients. (A) Sex; (B) Age; (C) Clinical stage; (D) Histological grade. Figure 5. ROC analysis for predicting prognosis. (A) Overall survival (OS); (B) Disease-free survival (DFS).
Figure 5. ROC analysis for predicting prognosis. (A) Overall survival (OS); (B) Disease-free survival (DFS). Figure 6. APOC1 expression analyses based on clinicopathological factors. (A) Sex; (B) Primary tumor laterality; (C) Age; (D) Histological grade; (E) T stage; (F) Clinical stage. Compared with normal group: ** P<0.01 and *** P<0.001.
Figure 6. APOC1 expression analyses based on clinicopathological factors. (A) Sex; (B) Primary tumor laterality; (C) Age; (D) Histological grade; (E) T stage; (F) Clinical stage. Compared with normal group: ** P<0.01 and *** P<0.001. Figure 7. The expression of co-expressed genes with APOC1 in various cells.
Figure 7. The expression of co-expressed genes with APOC1 in various cells. Figure 8. GO annotation and KEGG pathway analyses. (A) Molecular function (MF); (B) Biological process (BP); (C) Cellular component (CC); (D) Biological pathways.
Figure 8. GO annotation and KEGG pathway analyses. (A) Molecular function (MF); (B) Biological process (BP); (C) Cellular component (CC); (D) Biological pathways. Tables
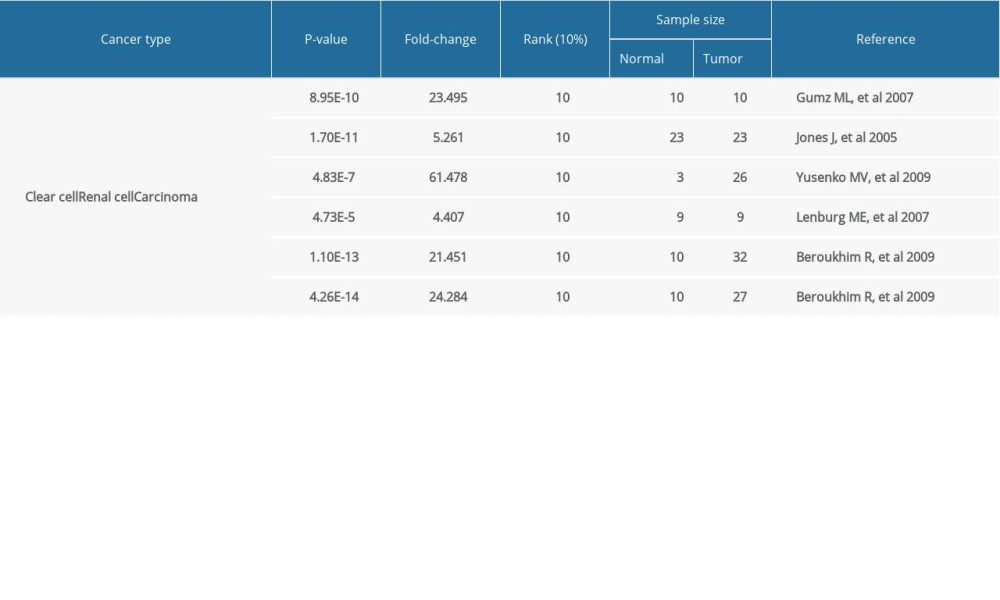 Table 1. APOC1 expression in clear cell renal cell carcinoma (cc RCC).
Table 1. APOC1 expression in clear cell renal cell carcinoma (cc RCC).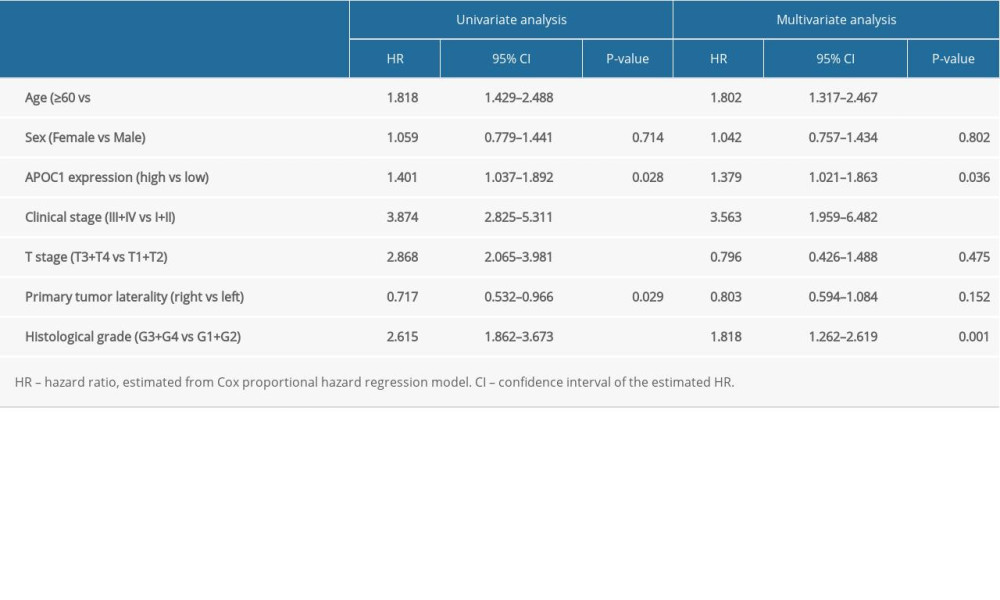 Table 2. Association of OS and clinical pathological characteristics in patients using Cox regression analysis.
Table 2. Association of OS and clinical pathological characteristics in patients using Cox regression analysis. Table 1. APOC1 expression in clear cell renal cell carcinoma (cc RCC).
Table 1. APOC1 expression in clear cell renal cell carcinoma (cc RCC). Table 2. Association of OS and clinical pathological characteristics in patients using Cox regression analysis.
Table 2. Association of OS and clinical pathological characteristics in patients using Cox regression analysis. In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








