27 December 2020: Database Analysis
Expression of the Topoisomerase II Alpha (TOP2A) Gene in Lung Adenocarcinoma Cells and the Association with Patient Outcomes
Xiaomei Du1AEFG, Zhiwen Xue1BF, Jianning Lv1BC, Heidou Wang2ACG*DOI: 10.12659/MSM.929120
Med Sci Monit 2020; 26:e929120
Abstract
BACKGROUND: This study was carried out to analyze TOP2A expression in lung adenocarcinoma (LUAD) and to assess its value in clinical diagnosis and prognosis.
MATERIAL AND METHODS: The Cancer Genome Atlas (TCGA) database was used to study the relationship of TOP2A expression with the progression and prognosis of LUAD. For a further elucidation of the value of TOP2A in LUAD, the effect of TOP2A knockout on cell viability and related protein expression of LUAD cell line A549 in vitro was investigated by using RNA interference, MTT, flow cytometry, RT-PCR, and western blot analysis.
RESULTS: According to the results of database analysis, TOP2A expression in LUAD was higher than that in normal lung tissues. There was a strong correlation of TOP2A expression with clinicopathological and epidemiological parameters of LUAD. The survival rate of LUAD patients with high TOP2A expression was lower than that of patients with low expression (P<0.001). The expression of TOP2A in A549 cells was higher than that in Beas-2B cells. After decreased expression of TOP2A in A549 cells, the proliferation of A549 cells was downregulated and the apoptosis rate was increased. It was further verified that TOP2A low expression exerts a role in LUAD through activation of the ERK/JNK/p-P38/CHOP signaling pathway.
CONCLUSIONS: The findings from this study showed that TOP2A expression was upregulated in a human lung adenocarcinoma cell line, and this finding was supported by bioinformatics analysis. Further studies are required to determine whether TOP2A expression is a prognostic biomarker and potential therapeutic target in patients with lung adenocarcinoma.
Keywords: database, Diagnosis, DNA Topoisomerases, Type II, Lung Diseases, A549 cells, Adenocarcinoma of Lung, Down-Regulation, Genetic Association Studies, RNA, Messenger
Background
Lung cancer has a high incidence rate and a high degree of malignancy [1]. Epidemiological data on lung cancer in China reveals that about 80% of lung cancers are non-small cell lung cancer, of which most are adenocarcinoma [2]. Lung adenocarcinoma (LUAD) has become one of the most important pathological subtypes of lung cancer and is an important research focus [3]. LUAD has an insidious morbidity, and two-thirds of patients are in advanced stage at the time of diagnosis [4]. The metastasis and poor prognosis of LUAD pose great challenges for the treatment of this type of cancer [5]. At present, there are various approaches available for the diagnosis of LUAD. However, there is a lack of adequate mature or ideal methods for its diagnosis. In addition, there is currently no effective diagnostic tool for the differential diagnosis of early and advanced LUAD.
The topoisomerase II alpha (TOP2A) gene, located on chromosome 17 (17q21-q22), encodes a 170-kDa DNA topoisomerase II α [6]. TOP2A is a cell cycle-dependent protein, whose expression is highly dependent on cell proliferation and can be suppressed when cell proliferation and division stop [7,8]. TOP2A is also essential for embryonic development in mammals. It has been confirmed that knockout of the mouse TOP2A gene can cause early death of embryos at cell stages 4–8 [9]. Significantly, TOP2A is essentially involved in various cell biological processes, such as DNA replication, chromatin condensation, chromosome separation, and structural maintenance of chromosomes [8,10–12]. Accordingly, multiple studies have been published to support that TOP2A gene expression is strongly associated with the occurrence and development of many tumors [13–17]. There have been reports focusing on the role of TOP2A gene in the development of LUAD [13,14]. For instance, Ma et al. proposed in their latest research that LUAD is the core gene in the LUAD-related gene network [13]. For a better understanding of the role of TOP2A in the occurrence and development of LUAD, the present study was conducted to analyze the expression of TOP2A in LUAD tissues and normal lung specimens using TCGA database and the Human Protein Atlas database, and to evaluate its value as a biomarker for diagnosis and prognosis of LUAD. With respect to the above, the present study was carried out to explore the expression of TOP2A in LUAD and its significance in clinical diagnosis and prognosis analysis on the using large-scale clinical data provided by bioinformatics network tools and the application of cell experiment techniques, so as to provide a theoretical basis for clinical diagnosis and drug targets in the future.
Material and Methods
GEPIA:
GEPIA is an interactive web server for cancer expression profile data developed by Peking University [18]. It is available to analyze the differential expression of genes from TCGA and GTEx and the corresponding correlation with prognosis. In view of the above, this study used GEPIA database to analyze the transcription level of TOP2A gene in LUAD tissues and normal lung tissues, as well as the OS of patients. GEPIA can adjust the queue threshold using the log-rank test, and the obtained survival plots also included Cox proportional hazard ratio and 95% confidence interval.
UALCAN:
We used the UALCAN database [19] to obtain data related to the expression of multiple genes in tumors and their direct relationship with prognosis. The UALCAN database includes GEx profile and Survival Profile, which can facilitate the analysis of gene expression as well as the relationship between gene expression and prognosis. Moreover, the UALCAN database can provide direct data on the expression of multiple genes in tumor tissue and the relationship with prognosis of patients. Therefore, in this study, the UALCAN database was used to detect the expression of TOP2A in LUAD patients with different pathological features and to analyze the survival of patients.
LINKEDOMICS:
LinkedOmics, which contains multiple sets of data for 32 cancer types from TCGA, has 3 analysis modules: LinkFinder, LinkInterpreter, and LinkCompare. In the present study, the LinkedOmics website was used to evaluate the association between the expression of TOP2AmRNA and the pathological features of LUAD patients, and to analyze its impact on the survival rate of patients. Non-parametric testing was used for statistical analysis.
KAPLAN-MEIER PLOTTER:
The Kaplan-Meier Plotter website is available for analyzing the mRNA expression of breast cancer, ovarian cancer, lung cancer, gastric cancer, and prognosis of breast cancer miRNA. It provides access to 18 674 cancer samples to facilitate the assessment of the impact of 54 675 genes on patient survival. In this study, online Kaplan-Meier Plotter survival analysis software was used to detect the effect of TOP2A gene on survival time of patients with LUAD.
THE HUMAN PROTEIN ATLAS (HPA):
The HPA database contains tissue and cellular distribution information of all 24 000 human proteins by using immunohistochemical technique with specific antibodies. It can eventually provide at least 576 immunohistochemical staining graphs, which are checked and indexed by professionals. In addition to facilitating immunostaining of tissues and cell lines, the HPA database can be used for differential expression analysis of proteins between normal lung tissues and tumor tissues. In this study, immunohistochemistry was performed to analyze the expression of TOP2A protein in normal lung tissues and LUAD tissues by using the immunohistochemical antibody of TOP2A(HPA023083) in the HPA database.
CELL CULTURE AND TRANSFECTION:
A549 and Beas-2B cell lines were purchased from the Typical Culture Preservation Commission Cell Bank, Chinese Academy of Science (Shanghai, China), which were cultured in DMEM medium containing fetal bovine serum (Hyclone, Logan, USA). After cell fusion reached about 80%, cells were digested with trypsin at room temperature. The cell liquid was then collected and centrifuged in a centrifuge tube for about 5 min, after which the supernatant was discarded. Subsequently, cells were added with 5 ml of culture medium, followed by a gentle blowing and beating to make the cells completely suspended, which were then inoculated in a new culture flask. The digested cells were then cultured in a cell incubator with constant temperature and pressure to make the cells adhere to the wall completely. TOP2AsiRNA was used to knock out TOP2A of A549 cells. The constructed plasmid was mixed with Lipofectamine3000 to transfect A549 cells for 48 h. TOP2A siRNA and negative control siRNA used in the experiment were purchased from GenePharma Company.
WESTERN BLOT:
Following total protein extraction from cells, protein quantification was carried out using BCA method. Afterwards, SDS-PAGE gel electrophoresis was performed on the target protein sample, with the condition of at 90 V for the concentration gel (upper) for 30 min, and at 120 V for the separation gel (lower) for 60 min. After membrane transfer, protein samples were them sealed in 50 g/L skimmed milk for 1 h. In the next step, the membrane was incubated with the primary antibody (Cell Signaling Technology, USA; dilution 1: 1000) overnight, followed by incubation with horseradish-peroxidase conjugated secondary IgG antibody (Cell Signaling Technology, Danvers, USA). After that, signals were observed with a chemiluminescence reagent (Millipore, Billerica, USA) and the protein bands were scanned using the ChemiDoc System (Bio-Rad). The main antibodies used in this experiment were provided by CST.
RT-PCR:
After total RNA extraction from the cultured cells using RNAisoPLUS, cDNA was reverse-transcribed with a PrimeScript RT kit (Thermo Fisher Scientific, Waltham, USA).
FLOW CYTOMETRY FOR CELL APOPTOSIS DETECTION:
Cells in logarithmic phase were inoculated in a 60-mm cell culture dish at a cell concentration of 1×106. After 24 h of culture in the incubator, the cells were taken out for cell apoptosis detection by using an AnnexinVPI kit (Keygen, China). Double-labeling of the collected cells was performed with AnnexinV and PI according to the instructions of the kit. Cells were then detected by flow cytometry after 30 min of staining.
STATISTICAL ANALYSIS:
All data analyses were performed using GraphPad Prism 8. The
Results
EXPRESSION OF TOP2A IN LUAD AND NORMAL LUNG TISSUES:
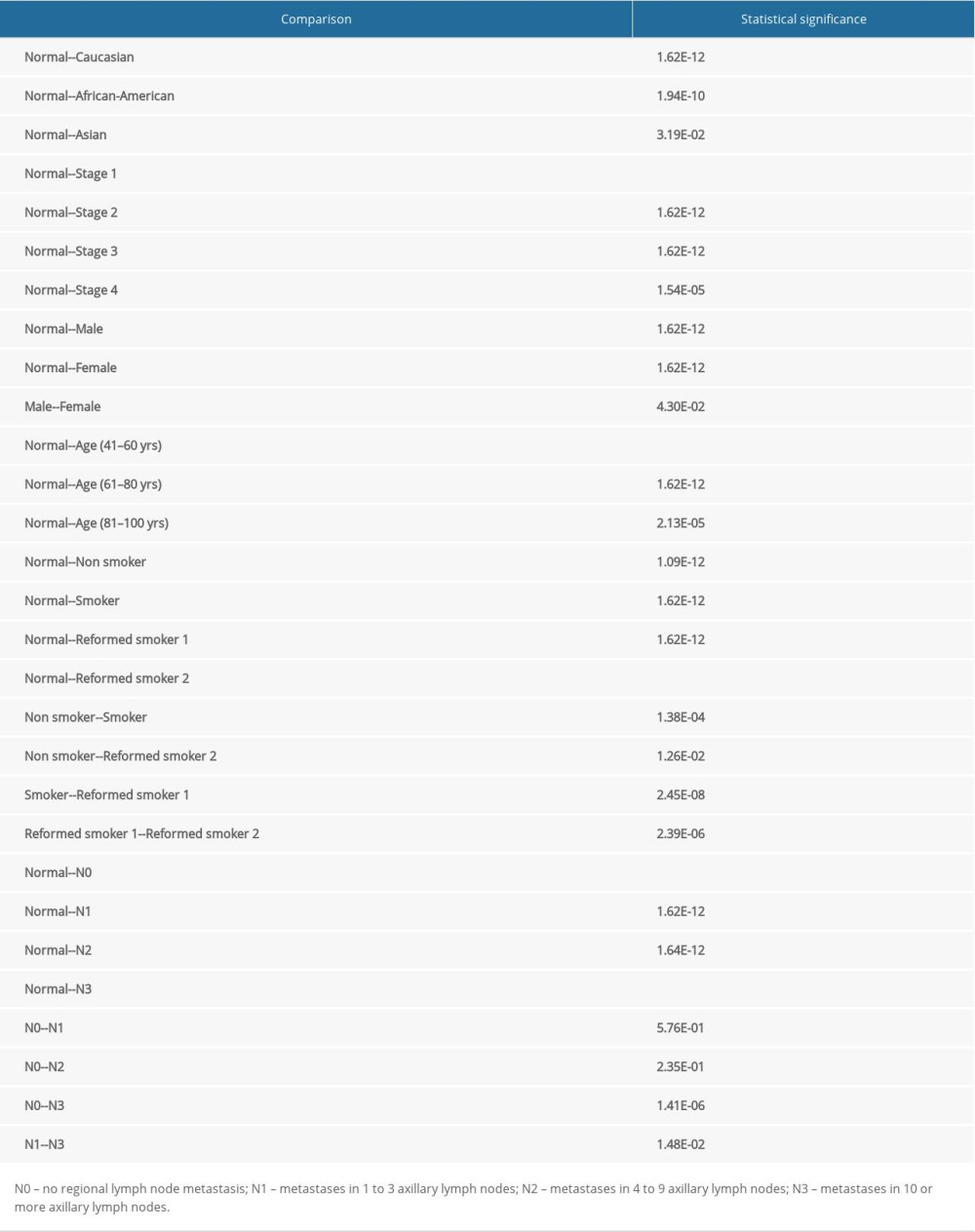
The expression of TOP2A mRNA in 483 normal lung tissues and 347 LUAD tumor specimens in TCGA database was detected in the GEPIA database, with the aim to explore the relationship between TOP2AmRNA expression and LUAD. The results showed that compared with normal lung tissues, the expression of TOP2A was obviously upregulated in LUAD (Figure 1A).
In addition, bioinformatic analysis was performed for TOP2A mRNA expression in 515 LUAD patients by using UALCAN. The results revealed that TOP2A mRNA expression was significantly increased in LUAD tissues when compared with the matched normal lung tissue (P<0.001) (Figure 1B).
To verify the results obtained from GEPIA and UALCAN, RT-PCR and western blot analyses were performed to detect the mRNA and protein expressions of TOP2A in Beas-2B and A549 cells. The results showed that the mRNA and protein expressions of TOP2A were significantly higher in A549 cells than those in Beas-2B cells. These results were consistent with the results of GEPIA and UALCAN (Figue 1C, 1D).
EXPRESSION OF TOP2A IN NORMAL LUNG TISSUE AND LUAD:
Immunohistochemistry was performed to analyze the protein expression of TOP2A in normal lung tissues and LUAD tissues by using the immunohistochemical antibody of TOP2A (HPA023083) in the HPA database. We found that the expression of TOP2A was lower in normal lung tissue than that in LUAD (Figure 2A).
Further survival analysis revealed that the survival rate of patients with high TOP2A expression was significantly lower than that of patients with low TOP2A expression (P<0.001, Figure 2B).
CORRELATION ANALYSIS BETWEEN TOP2A MRNA AND PATHOLOGICAL FEATURES OF LUAD USING UALCAN AND LINKEDOMICS DATABASE:
According to the results of relationship analysis between TOP2A mRNA expression and pathological features of LUAD using UALCAN database, TOP2A mRNA expression was associated with age, sex, race, individual cancer stages, and nodal metastasis status (Figure 3A–3F). Table 1 lists the specific results of P values.
The LinkedOmics database was also utilized to analyze the relationship between TOP2AmRNA expression and pathological features of LUAD. As indicated by the results, TOP2A mRNA expression exhibited an association with radiation therapy (P=1.109e-02, n=469), histological type (P=1.262e-06, n=515), pathologic stage (P=1.607e-02, n=507), T stage (P=1.721e-03, n=512), ethnicity (P=1.947e-02, n=390), and N stage (P=5.798e-02, n=503). The above results suggest that TOP2A mRNA is strongly associated with the occurrence and development of LUAD (Figure 4).
TOP2A MRNA AS AN INDEPENDENT PROGNOSTIC FACTOR FOR LUAD:
In accordance with the high correlation between TOP2A mRNA expression and the progression of LUAD, we speculated whether TOP2A mRNA expression can be used as an independent prognostic indicator in patients with LUAD. The Kaplan-Meier Plotter database was used to assess the association of TOP2A mRNA expression with the OS of patients. Corresponding results that the OS of patients with high TOP2A mRNA expression was significantly shorter than that of patients with low expression [log-rank P<1e-16, HR=2 (1.78–2.23); Figure 5A].
GEPIA was further applied to analyze the relationship of TOP2A mRNA expression with the OS of patients. It was discovered that patients with high expression of TOP2A mRNA had obviously lower OS than that of patients with low expression (log-rank P=0.011, HR=1.5, Figure 5B).
To further clarify the role of TOP2A in LUAD, 492 cases of LUAD were analyzed using the LinkedOmics database. The survival time of patients with high TOP2A mRNA expression was remarkably shorter than that of patients with low expression, and the difference was statistically significant (P=2.380e-03, Figure 5C).
ROLE OF TOP2A IN LUAD VIA MAPK SIGNALING PATHWAY:
For further elucidation of the effect of TOP2A in LUAD, TOP2A in A549 cells was knocked out with RNA interference technology, and siTOP2A cells were detected by using RT-PCR and western blot. The results revealed that TOP2A gene expression was obviously reduced in siTOP2A cells (Figure 6). In addition, MTT was applied to detect cell viability and cell proliferation in siTOP2A and siControl cells. It was found that the cell viability of siA549 cells decreased after decreased expression of TOP2A (Figure 6). Meanwhile, cell apoptosis detection by using flow cytometry indicated that the apoptosis rate in siTOP2A was evidently higher than that in siControl (P<0.05, Figure 6). For a further clarification of the mechanism of TOP2A, the expression of MAPK in siTOP2A was detected by western blot, without obvious statistical significance in the total protein expression (P>0.05). We found a significant increase in the protein expression of p-ERK, p-JNK, p-P38, and CHOP in siTOP2A cells (P<0.05). The aforementioned results suggest that TOP2A plays a role in promoting the development of LUAD by regulating the ERK/JNK/p-P38/CHOP signaling pathway.
Discussion
Lung cancer ranks first in incidence of all kinds of malignant tumors [20]. Current effective treatments of lung cancer include surgical resection, radiotherapy, and targeted therapy [2,21]. LUAD is the most common type of lung cancer, which is characterized by occult onset, extremely high incidence, and high risk of metastasis [2,3,22]. Due to the lack of an effective approach for early diagnosis, most patients have developed local infiltration or distant metastasis at diagnosis and thus have lost the chance to benefit from surgery [2,4]. Radiotherapy and chemotherapy are the major choices for patients with advanced LUAD, which accounts for about 40% of confirmed cases [21]. However, it has unsatisfactory treatment outcomes, with a poor 5-year survival rate of only 5–10% [21,23]. Therefore, the analysis of prognostic factors and screening of sensitive diagnostic markers for LUAD in the early stage are important topics in clinical research on LUAD. The findings of the present study support that TOP2A can be considered a newly discovered biomarker for the diagnosis, prognosis, and treatment of LUAD.
In the present study, for the exploration of the relationship between TOP2A and LUAD, GEPIA and UALCAN databases were used to detect the expression of TOP2A mRNA in normal lung tissues and LUAD tissues in TCGA database. Results of both databases showed that TOP2A expression in LUAD was significantly higher than that in normal lung tissue. In order to further verify the expression of TOP2A in LUAD, an
Substantial evidence supports that TOP2A functions significantly in DNA replication, transcription, recombination, and chromatin reconstruction [24]. It has been reported that cell division stops when the protein expression of TOP2A is silenced in human cells [25]. Accordingly, TOP2A is a cell proliferation-related protein, whose expression promotes the completion of cell division [11,17]. TOP2A gene expression has been found to be related to cell mitosis in S phase in breast cancer [7,26,27], and there is significant expression of TOP2A in different subtypes of breast cancer. Specifically, the protein expression of TOP2A is high in high-proliferative subtypes of breast cancer (e.g., basal-like, Luminal B, and HER2-positive) [28]. In this regard, TOP2A can be used as a tumor marker of breast cancer [28]. Furthermore, high expression of TOP2A protein was reported to be associated with malignant tumor proliferation and high histological grade of breast cancer, suggesting a poor prognosis in breast cancer patients with high TOP2A gene expression [29]. These results suggest that TOP2A protein can be considered as a molecular target for the treatment of breast cancer, and can also be a prognostic factor. Basic research revealed that the knockdown of TOP2A can retard cell proliferation and suppress tumor formation [25]. It has also been proposed that TOP2A is a core protease that promotes and maintains tumor progression in tumor cells [13,17,25]. In assessing the role of the TOP2A gene, most studies focused on breast cancer, gastric cancer, liver cancer, prostate cancer, colorectal cancer, and esophageal cancer [6,27,30–33]. Increasingly more studies have been reported focusing on the effect of TOP2A gene on the clinical development and prognosis of lung adenocarcinoma [13,14]. For example, Fan Kou et al. demonstrated in their research that TOP2A could promote lung adenocarcinoma cells’ malignant progression and predict poor prognosis in lung adenocarcinoma [14]. This is consistent with the results in our study, revealing that the survival time of patients with high TOP2A mRNA expression was significantly shorter than that of patients with low TOP2A mRNA expression.
Collectively, the findings in our study support that TOP2A could be a new biomarker for diagnosis, prognosis, and treatment of LUAD. However, our findings are insufficient to establish that TOP2A can be used as an indicator for clinical diagnosis and prognosis in the clinical setting. Accordingly, there is a need to carry out more clinical trials and studies for further verification.
Conclusions
The findings from this study show that TOP2A expression is upregulated in a human lung adenocarcinoma cell line, and this finding was supported by bioinformatics analysis. Further studies are required to determine whether TOP2A expression is a prognostic biomarker and potential therapeutic target in patients with adenocarcinoma of the lung.
Figures
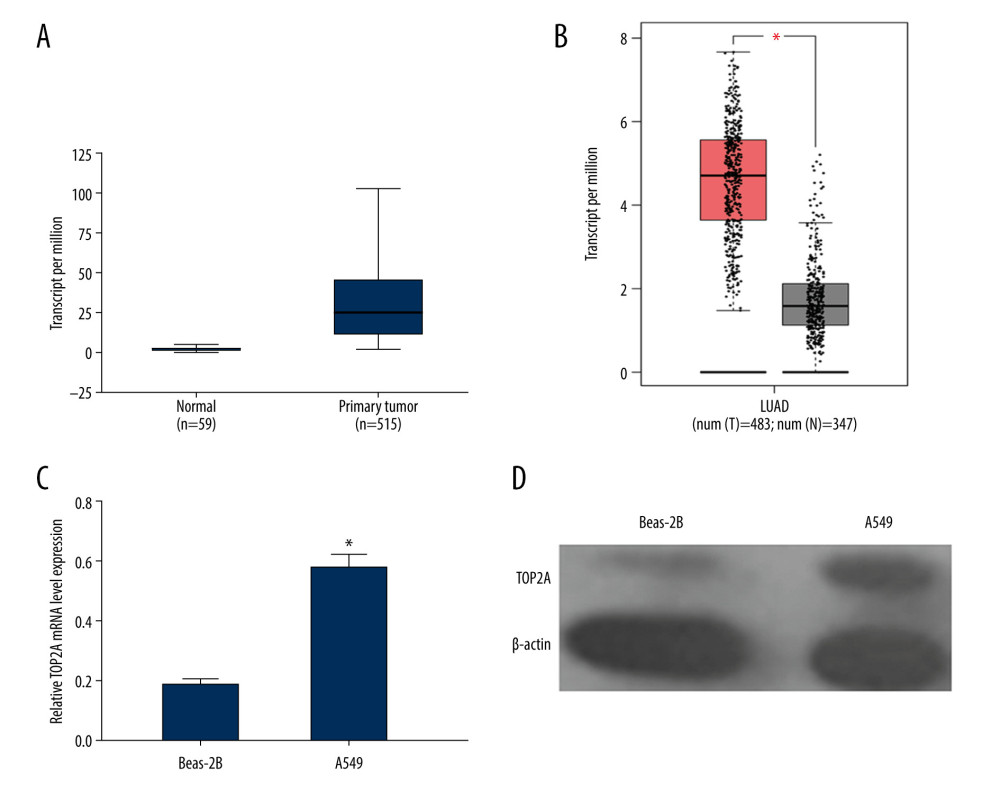 Figure 1. TOP2A mRNA is downregulated in LUAD used TCGA database. (A) Box plots and P values was produced using UALCAN. (B) Box plots and P values was produced using GEPIA. (C) mRNA expression of TOP2A in Beas-2B and A549 cells. (D) Protein expression of TOP2A in Beas-2B and A549 cells.
Figure 1. TOP2A mRNA is downregulated in LUAD used TCGA database. (A) Box plots and P values was produced using UALCAN. (B) Box plots and P values was produced using GEPIA. (C) mRNA expression of TOP2A in Beas-2B and A549 cells. (D) Protein expression of TOP2A in Beas-2B and A549 cells. 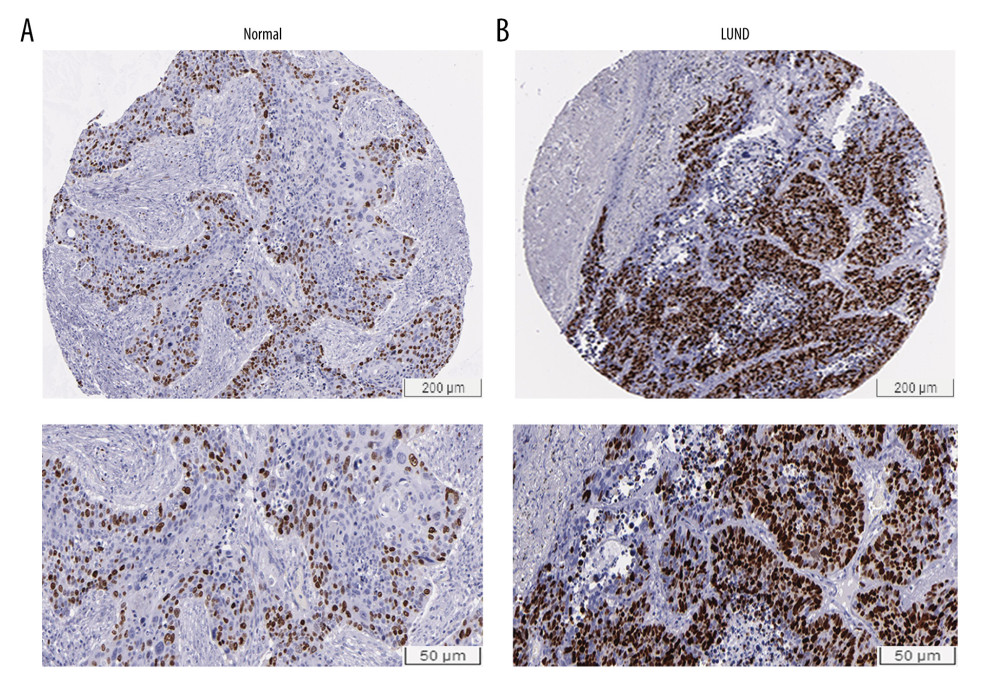 Figure 2. Immunohistochemical examination for expression of TOP2A in normal lung and LUAD tissue. (A) TOP2A expression in normal lung tissues. (B) TOP2A expressions in LUAD tissues.
Figure 2. Immunohistochemical examination for expression of TOP2A in normal lung and LUAD tissue. (A) TOP2A expression in normal lung tissues. (B) TOP2A expressions in LUAD tissues.  Figure 3. Association of TOP2A mRNA and pathological features in LUAD. (A) TOP2A mRNA with age (B). TOP2A mRNA with gender. (C) TOP2A mRNA with individual cancer stages. (D) TOP2A mRNA with race. (E). TOP2A mRNA with patient’s smoking habits. (F) TOP2A mRNA with nodal metastasis status. Box plots and P values (Table 1) of A–F were produced using UALCAN.
Figure 3. Association of TOP2A mRNA and pathological features in LUAD. (A) TOP2A mRNA with age (B). TOP2A mRNA with gender. (C) TOP2A mRNA with individual cancer stages. (D) TOP2A mRNA with race. (E). TOP2A mRNA with patient’s smoking habits. (F) TOP2A mRNA with nodal metastasis status. Box plots and P values (Table 1) of A–F were produced using UALCAN. 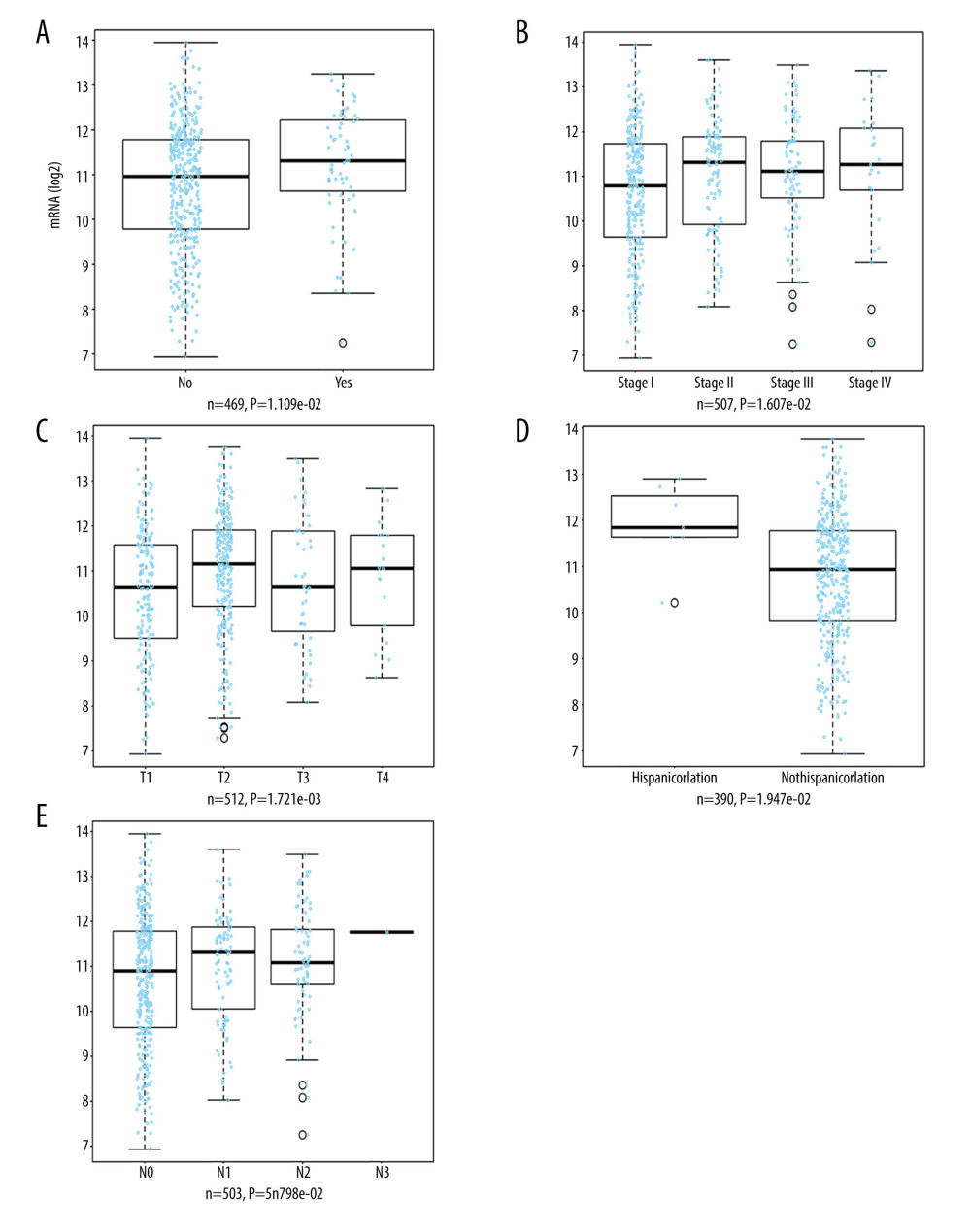 Figure 4. LinkedOmics analysis for the relationship between TOP2A mRNA expression level and clinicopathological features of LUAD. (A) Radiation therapy; (B) pathologic stage; (C) pathology t stage; (D) ethnicity; (E) pathology n stage. Box plots and P values of were produced using LinkedOmics.
Figure 4. LinkedOmics analysis for the relationship between TOP2A mRNA expression level and clinicopathological features of LUAD. (A) Radiation therapy; (B) pathologic stage; (C) pathology t stage; (D) ethnicity; (E) pathology n stage. Box plots and P values of were produced using LinkedOmics. 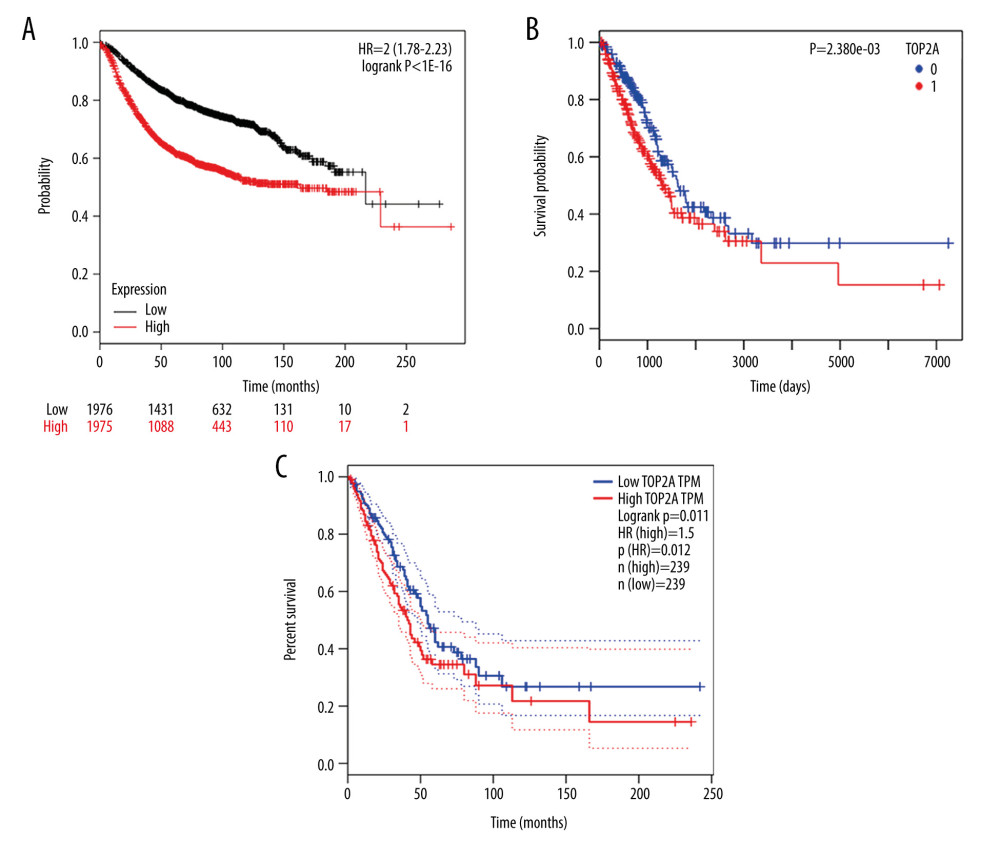 Figure 5. TOP2A mRNA is an independent favorable prognostic factor for LUAD. (A) Kaplan-Meier plot was produced using Kaplan-Meier Plotter. (B) Kaplan-Meier plot was produced using LinkedOmics. (C) The survival rate was produced using GEPIA.
Figure 5. TOP2A mRNA is an independent favorable prognostic factor for LUAD. (A) Kaplan-Meier plot was produced using Kaplan-Meier Plotter. (B) Kaplan-Meier plot was produced using LinkedOmics. (C) The survival rate was produced using GEPIA. 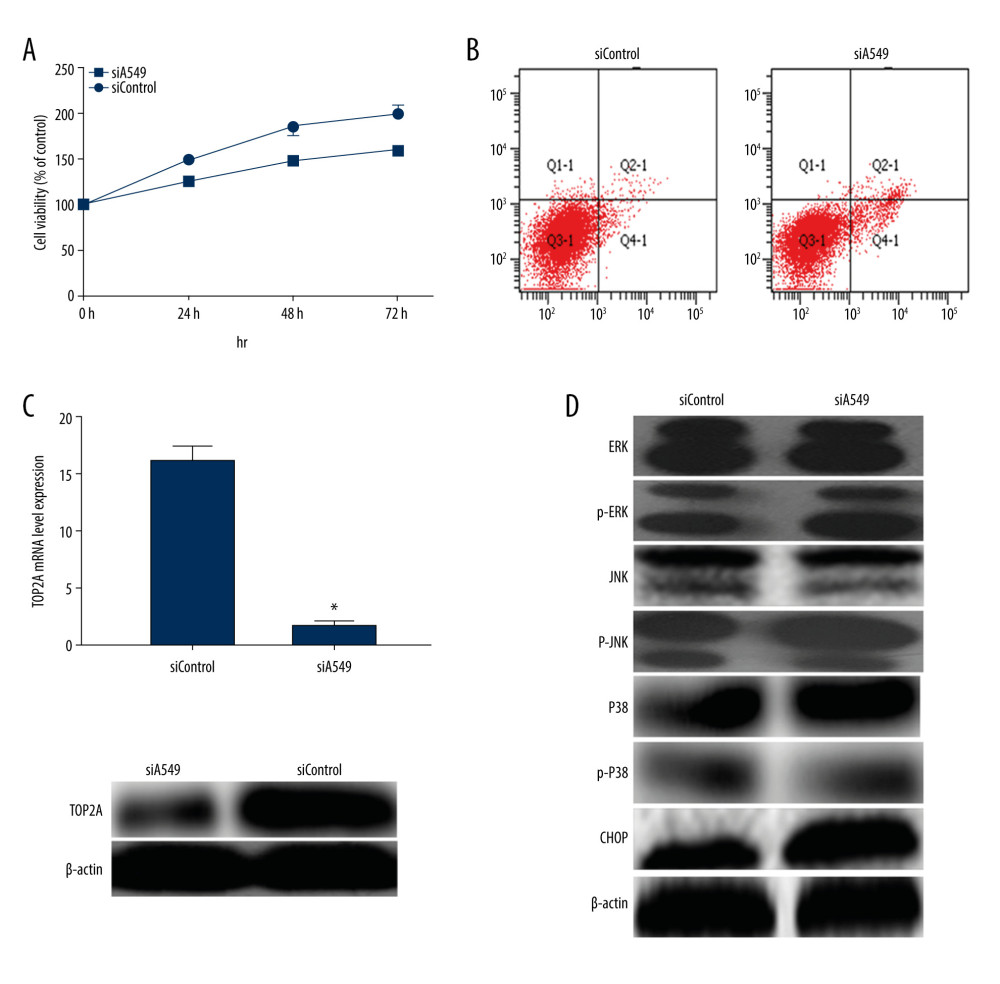 Figure 6. Measurement of relevant indexes after low expression of TOP2A in A549 cells. (A) Cell viability detection by MTT assay. Cell viability of siA549 was significantly lower than that of siControl (* P<0.01). (B) Cell apoptosis detection by flow cytometry. Cell apoptosis of siA549 was obviously higher than that of siControl (* P<0.01). (C) Detection of TOP2A mRNA and protein expressions in siA549 and siControl cells by RT-PCR and Western blot. (D) Detection of proteins related to ERK/JNK/p-P38/CHOP signaling pathway using Western blot.
Figure 6. Measurement of relevant indexes after low expression of TOP2A in A549 cells. (A) Cell viability detection by MTT assay. Cell viability of siA549 was significantly lower than that of siControl (* P<0.01). (B) Cell apoptosis detection by flow cytometry. Cell apoptosis of siA549 was obviously higher than that of siControl (* P<0.01). (C) Detection of TOP2A mRNA and protein expressions in siA549 and siControl cells by RT-PCR and Western blot. (D) Detection of proteins related to ERK/JNK/p-P38/CHOP signaling pathway using Western blot. References
1. Dela Cruz CS, Tanoue LT, Matthay RA, Lung cancer: Epidemiology, etiology, and prevention: Clin Chest Med, 2011; 32(4); 605-44
2. Molina JR, Yang P, Cassivi SD, Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship: Mayo Clinic Proc, 2008; 83(5); 584-94
3. Butnor KJ, Controversies and challenges in the histologic subtyping of lung adenocarcinoma: Transl Lung Cancer Res, 2020; 9(3); 839-46
4. Myers DJ, Wallen JM, Cancer, lung adenocarcinoma. [Updated 2020 Jun 26]: StatPearls, 2020, Treasure Island (FL), StatPearls Publishing https://www.ncbi.nlm.nih.gov/books/NBK519578/
5. Kamath SD, Kumthekar PU, Kruser TJ, Mohindra NA, Intracranial response to anti-programmed death 1 therapy in a patient with metastatic non-small cell lung cancer with leptomeningealcarcinomatosis: Oncologist, 2018; 23(12); e159-61
6. Panvichian R, Tantiwetrueangdet A, Angkathunyakul N, Leelaudomlipi S, TOP2A amplification and overexpression in hepatocellular carcinoma tissues: Biomed Res Int, 2015; 2015 381602
7. Chen T, Sun Y, Ji P, Topoisomerase IIα in chromosome instability and personalized cancer therapy: Oncogene, 2015; 34(31); 4019-31
8. Wang JC, Cellular roles of DNA topoisomerases: A molecular perspective: Nat Rev Mol Cell Biol, 2002; 3(6); 430-40
9. Sapetto-Rebow B, McLoughlin SC, O’Shea LC, Maternal topoisomerase II alpha, not topoisomerase II beta, enables embryonic development of zebrafish top2a−/− mutants: BMC Dev Biol, 2011; 11; 71
10. Thakurela S, Garding A, Jung J, Gene regulation and priming by topoisomerase IIα in embryonic stem cells: Nat Commun, 2013; 4; 2478
11. Nitiss JL, DNA topoisomerase II and its growing repertoire of biological functions: Nat Rev Cancer, 2009; 9(5); 327-37
12. Morimoto S, Tsuda M, Bunch H, Type II DNA topoisomerases cause spontaneous double-strand breaks in genomic DNA: Genes, 2019; 10(11); 868
13. Ma W, Wang B, Zhang Y, Prognostic significance of TOP2A in non-small cell lung cancer revealed by bioinformatic analysis: Cancer Cell Int, 2019; 9; 239
14. Kou F, Sun H, Wu L, TOP2A promotes lung adenocarcinoma cells’ malignant progression and predicts poor prognosis in lung adenocarcinoma: J Cancer, 2020; 11(9); 2496-508
15. Hooks KB, Audoux J, Fazli H, New insights into diagnosis and therapeutic options for proliferative hepatoblastoma: Hepatology, 2018; 68(1); 89-102
16. Heestand GM, Schwaederle M, Gatalica Z, Topoisomerase expression and amplification in solid tumours: Analysis of 24,262 patients: Eur J Cancer, 2017; 83; 80-87
17. Kou F, Sun H, Wu L, TOP2A promotes lung adenocarcinoma cells’ malignant progression and predicts poor prognosis in lung adenocarcinoma: J Cancer, 2020; 11(9); 2496-508
18. Feng Y, Guo C, Wang H, Fibrinogen-like protein 2 (FGL2) is a novel biomarker for clinical prediction of human breast cancer: Med Sci Monit, 2020; 26; e923531
19. Zhou Z, Wu B, Tang X, High SET somain Bifurcated 1 (SETDB1) expression predicts poor prognosis in breast carcinoma: Med Sci Monit, 2020; 26; e922982
20. Barta JA, Powell CA, Wisnivesky JP, Global epidemiology of lung cancer: Ann Glob Health, 2019; 85(1); 8
21. Zappa C, Mousa SA, Non-small cell lung cancer: Current treatment and future advances: Transl Lung Cancer Res, 2016; 5(3); 288-300
22. Alberg AJ, Brock MV, Ford JG: Chest, 2013; 143(5 Suppl); e1S-29S
23. Lu T, Yang X, Huang Y, Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades: Cancer Manag Res, 2019; 11; 943-53
24. Yu X, Davenport JW, Urtishak KA: Genome Res, 2017; 27(7); 1238-49
25. Zhang R, Xu J, Zhao J, Bai JH, Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A: J Cell Biochem, 2018; 119(9); 7256-63
26. Kang X, Song C, Du X, PTEN stabilizes TOP2A and regulates the DNA decatenation: Sci Rep, 2015; 5; 17873
27. Jain M, Zhang L, He M, TOP2A is overexpressed and is a therapeutic target for adrenocortical carcinoma: Endocr Relat Cancer, 2013; 20(3); 361-70
28. Romero A, Martín M, Cheang MC: Am J Pathol, 2011; 178(4); 1453-60
29. Ren L, Liu J, Gou K, Xing C, Copy number variation and high expression of DNA topoisomerase II alpha predict worse prognosis of cancer: A meta-analysis: J Cancer, 2018; 9(12); 2082-92
30. Coss A, Tosetto M, Fox EJ, Increased topoisomerase IIalpha expression in colorectal cancer is associated with advanced disease and chemotherapeutic resistance via inhibition of apoptosis: Cancer Lett, 2009; 276(2); 228-38
31. Schaefer-Klein JL, Murphy SJ, Johnson SH, Topoisomerase 2 alpha cooperates with androgen receptor to contribute to prostate cancer progression: PLoS One, 2015; 10(11); e0142327
32. Liu LM, Xiong DD, Lin P, DNA topoisomerase 1 and 2A function as oncogenes in liver cancer and may be direct targets of nitidine chloride: Int J Oncol, 2018; 53(5); 1897-912
33. Kanta SY, Yamane T, Dobashi Y: Hum Pathol, 2006; 37(10); 1333-43
Figures
 Figure 1. TOP2A mRNA is downregulated in LUAD used TCGA database. (A) Box plots and P values was produced using UALCAN. (B) Box plots and P values was produced using GEPIA. (C) mRNA expression of TOP2A in Beas-2B and A549 cells. (D) Protein expression of TOP2A in Beas-2B and A549 cells.
Figure 1. TOP2A mRNA is downregulated in LUAD used TCGA database. (A) Box plots and P values was produced using UALCAN. (B) Box plots and P values was produced using GEPIA. (C) mRNA expression of TOP2A in Beas-2B and A549 cells. (D) Protein expression of TOP2A in Beas-2B and A549 cells. Figure 2. Immunohistochemical examination for expression of TOP2A in normal lung and LUAD tissue. (A) TOP2A expression in normal lung tissues. (B) TOP2A expressions in LUAD tissues.
Figure 2. Immunohistochemical examination for expression of TOP2A in normal lung and LUAD tissue. (A) TOP2A expression in normal lung tissues. (B) TOP2A expressions in LUAD tissues. Figure 3. Association of TOP2A mRNA and pathological features in LUAD. (A) TOP2A mRNA with age (B). TOP2A mRNA with gender. (C) TOP2A mRNA with individual cancer stages. (D) TOP2A mRNA with race. (E). TOP2A mRNA with patient’s smoking habits. (F) TOP2A mRNA with nodal metastasis status. Box plots and P values (Table 1) of A–F were produced using UALCAN.
Figure 3. Association of TOP2A mRNA and pathological features in LUAD. (A) TOP2A mRNA with age (B). TOP2A mRNA with gender. (C) TOP2A mRNA with individual cancer stages. (D) TOP2A mRNA with race. (E). TOP2A mRNA with patient’s smoking habits. (F) TOP2A mRNA with nodal metastasis status. Box plots and P values (Table 1) of A–F were produced using UALCAN. Figure 4. LinkedOmics analysis for the relationship between TOP2A mRNA expression level and clinicopathological features of LUAD. (A) Radiation therapy; (B) pathologic stage; (C) pathology t stage; (D) ethnicity; (E) pathology n stage. Box plots and P values of were produced using LinkedOmics.
Figure 4. LinkedOmics analysis for the relationship between TOP2A mRNA expression level and clinicopathological features of LUAD. (A) Radiation therapy; (B) pathologic stage; (C) pathology t stage; (D) ethnicity; (E) pathology n stage. Box plots and P values of were produced using LinkedOmics. Figure 5. TOP2A mRNA is an independent favorable prognostic factor for LUAD. (A) Kaplan-Meier plot was produced using Kaplan-Meier Plotter. (B) Kaplan-Meier plot was produced using LinkedOmics. (C) The survival rate was produced using GEPIA.
Figure 5. TOP2A mRNA is an independent favorable prognostic factor for LUAD. (A) Kaplan-Meier plot was produced using Kaplan-Meier Plotter. (B) Kaplan-Meier plot was produced using LinkedOmics. (C) The survival rate was produced using GEPIA. Figure 6. Measurement of relevant indexes after low expression of TOP2A in A549 cells. (A) Cell viability detection by MTT assay. Cell viability of siA549 was significantly lower than that of siControl (* P<0.01). (B) Cell apoptosis detection by flow cytometry. Cell apoptosis of siA549 was obviously higher than that of siControl (* P<0.01). (C) Detection of TOP2A mRNA and protein expressions in siA549 and siControl cells by RT-PCR and Western blot. (D) Detection of proteins related to ERK/JNK/p-P38/CHOP signaling pathway using Western blot.
Figure 6. Measurement of relevant indexes after low expression of TOP2A in A549 cells. (A) Cell viability detection by MTT assay. Cell viability of siA549 was significantly lower than that of siControl (* P<0.01). (B) Cell apoptosis detection by flow cytometry. Cell apoptosis of siA549 was obviously higher than that of siControl (* P<0.01). (C) Detection of TOP2A mRNA and protein expressions in siA549 and siControl cells by RT-PCR and Western blot. (D) Detection of proteins related to ERK/JNK/p-P38/CHOP signaling pathway using Western blot. In Press
05 Mar 2024 : Clinical Research
Effects of Thermal Insulation on Recovery and Comfort of Patients Undergoing Holmium Laser LithotripsyMed Sci Monit In Press; DOI: 10.12659/MSM.942836
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952