10 April 2021: Clinical Research
Clinical Application of Chromosomal Microarray Analysis in Pregnant Women with Advanced Maternal Age and Fetuses with Ultrasonographic Soft Markers
Zhu-Ming Hu1ABCE, Lei-Lei Li1ABD, Han Zhang1BF, Hong-Guo Zhang1BC, Rui-Zhi Liu1AFG, Yang Yu1ACF*DOI: 10.12659/MSM.929074
Med Sci Monit 2021; 27:e929074
Abstract
BACKGROUND: In pregnant women with advanced maternal age (AMA) and fetuses with ultrasonographic (USG) soft markers it is always challenging to decide whether to implement chromosomal microarray analysis (CMA) or not. It is unclear whether CMA should be used in the fetuses with isolated USG soft markers, and there is still a lack of extensive sample research.
MATERIAL AND METHODS: We enrolled 1521 cases in our research and divided them into 3 groups as follows: pregnant women with isolated AMA (group 1, n=633), pregnant women whose fetuses had isolated USG soft markers (group 2, n=750), and pregnant women with AMA whose fetuses had isolated USG soft markers (group 3, n=138). All pregnant women underwent prenatal ultrasound and amniocentesis, and fetal cells in the amniotic fluid were used for genetic analysis of CMA. All participants signed a written informed consent prior to CMA.
RESULTS: Abnormal findings were detected by CMA in 330 (21.70%) fetuses, including 37 (2.43%) clinically significant copy number variations (CNVs), 52 (3.42%) benign or likely benign CNVs, and 240 (15.78%) variants of unknown significance. The frequency of clinically significant CNVs in group 1 and group 2 were significantly lower than that in group 3 (2.37% and 2.0% vs 5.07%, P<0.01). More than a half (59.46%, 22/37) of the pregnant women decided to continue their pregnancy despite having a fetus diagnosed with clinically significant CNV.
CONCLUSIONS: CMA can increase the diagnostic yield of fetal chromosomal abnormality for pregnant women with isolated AMA or/and their fetuses had isolated USG soft markers.
Keywords: Chromosome Disorders, DNA Copy Number Variations, Karyotyping, Maternal Age, Prenatal Diagnosis, Aging, Amniocentesis, Chromosome Aberrations, Chromosomes, Human, Fetus, Microarray Analysis, Pregnancy, Ultrasonography
Background
Pregnant women with advanced maternal age (AMA) tend to have a high risk of trisomy 21, and it remains a common high-risk factor for fetal chromosomal aberrations [1]. In the past several decades, AMA has been an independent clinical indication of invasive prenatal diagnosis. For the past few years, the proportion of AMA pregnant woman has increased rapidly because of the full implementation of the universal two-child policy in China [2]. Abnormal chromosome numbers and the majority of structural chromosomal aberrations can be identified by conventional karyotyping. However, there are still many chromosomal abnormalities, such as chromosomal microdeletion and microduplication, which were undetectable by conventional karyotyping because of its limitation of low resolution [3–5].
Research shows that the proportion of clinically significant submicroscopic copy number variants (CNVs) detected by chromosomal microarray analysis (CMA) in fetuses with normal karyotype and abnormal ultrasonic appearance was 5.2–10% [6]. Wapner et al have reported that CMA can identify with approximately 6% of pregnancies with submicroscopic chromosomal abnormalities in the fetal with structural ultrasound abnormality and a normal karyotype [7]. However, fetal USG soft markers, commonly with no or little pathological clinical significance, are more often found in fetuses and are closely related to submicroscopic chromosomal imbalances [8]. Previous studies have confirmed that CMA can improve the detection rate of chromosomal submicroscopic aberrations for fetuses with an increased nuchal translucency (NT) and a normal karyotype. Therefore, it is suggested that increased NT should be recommended as an indication for CMA when the prenatal diagnosis is performed [9,10].
AMA pregnant women with or without fetal USG soft markers and the fetuses with normal karyotype are always hesitant when choosing whether to perform CMA. It is unclear whether CMA should be used in the fetus with a normal karyotype and isolated USG soft markers, and there is still a lack of large-sample research. The purpose of the present research was to estimate the application value of CMA in the prenatal diagnosis of AMA pregnant women with or without fetal USG soft markers and isolated fetal USG soft markers when the fetuses have a normal karyotype, and to provide further evidence for clinical genetic counseling practice.
Material and Methods
SUBJECTS:
We collected information on 2373 singleton-pregnant women with AMA (≥35 years) or/and ultrasound abnormalities who underwent amniocentesis and performed CMA from August 2016 to February 2020. However, 664 pregnant women with other high-risk factors of prenatal diagnosis besides AMA and USG soft markers were excluded, including adverse pregnancy history, familial chromosomal structural abnormality, screening indicating high risk of Down syndrome, fetal ultrasonic structural abnormality, or multiple high-risk factors listed here. We then excluded a further 188 pregnant women whose fetuses had an abnormal karyotype. As a result, a total of 1521 cases were included in this retrospective study. The mean age of the pregnant women was 33.27±5.57 years, and the mean gestational age when amniocentesis was performed was 141.63±19.86 days. Amniocentesis was performed during gestational weeks 18 and 32. Clinical genetic counseling was provided to all participants, including the fact that amniocentesis carries a risk of miscarriage (0.5~1%), the comparative advantages and limitations of traditional karyotype and CMA, respectively, and the possibility of detecting a variant of unknown significance (VUS).
We divided these 1521 cases into 3 groups: pregnant women with isolated AMA were included in group 1 (n=633), pregnant women whose fetuses had isolated USG soft markers were included in group 2 (n=750), and pregnant women with AMA and their fetuses that had isolated USG soft markers were included in group 3 (n=138). The USG soft markers included increased NT or nuchal fold (NF), cerebral ventriculomegaly, pyelic separation, echogenic intracardiac focus in the ventricle, intestinal echo enhancement, choroidplexuscyst, ductus venosus A wave inversion, nasal bone dysplasia, long bone dysplasias, and cervical lymphatic hygroma.
ETHICS APPROVAL:
The implementation of this research was consistent with ethics standards. The ‘Methods for Ethical Review of Biomedical Research Involving People’ issued by the relevant Chinese authorities and the ‘Declaration of Helsinki’ are the foundation of ethics standards. The Medical Ethics Committee of the First Hospital of Jilin University approved this study in 2016 (No. 2016-368). All participants signed a written informed consent prior to CMA.
CYTOGENETIC EXAMINATION:
At least 20 G-banded metaphases should be used for karyotype analysis in accordance with the operating manual of our center’s cytogenetics laboratory. Cultured amniotic fluid was arrested in metaphase, and G-banding was performed using Giemsa stain at a resolution of 320–400 bands. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN 2013) [11].
CHROMOSOMAL MICROARRAY ANALYSIS:
About 10 ml of amniotic fluid was collected by ultrasound-guided amniocentesis for CMA. Genomic DNA was extracted through the DNeasy Blood & Tissue kits (Qiagen GmBH, Hilden, Germany) in accordance with the manufacturer’s instructions. The NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used for the quantification of DNA. The Affymetrix CytoScan750K_Array (Affymetrix, Santa Clara, CA, USA) was used to detect the potential CNVs. DNA processing included digestion, joining, breaking, marking, hybridization, staining, and scanning. The data were analyzed using the software of the chromosome analysis suite (ChAS). The data on chromosomal microarray and genotype–phenotype correlations were analyzed by using the databases of Genomic Variants (
The types of CNVs clinical manifestations are: pathogenic, likely pathogenic, benign, likely benign, and variants of unknown significance (VUS). In addition, pathogenic and likely pathogenic CNVs are considered as clinically significant CNVs [12]. The types of CNVs clinical manifestations refers to the American College of Medical Genetics (ACMG) guidelines.
STATISTICAL ANALYSIS:
SPSS software v23.0 was used for statistical analysis of the data. An independent-sample
Results
PATIENT CLINICAL CHARACTERISTICS AND MAJOR CMA RESULTS:
In total, 1521 cases with normal karyotypes of fetuses were enrolled in this research and no more significant clinical findings were found except for the AMA and USG soft markers. The demographic data are shown in Table 1. The maternal age and gestational age at amniocentesis were significantly different (P<0.01) between the groups according to the independent-sample t test. There was no statistically significant difference (P>0.05) between the groups in pregnancy outcomes. No pregnant woman in this study had miscarriage due to amniocentesis.
Among the 1521 cases, abnormal CMA findings were detected in 330 (21.70%, 330/1521) fetuses, including 37 (2.43%, 37/1521) clinically significant CNVs, 52 (3.42%, 52/1521) benign or likely benign CNVs, 240 (15.78%, 240/1521) VUS, and 1 low-level mosaicism (14% ratio) of trisomy 15 (0.07%, 1/1521).
CMA FINDINGS FOR THE 3 GROUPS:
In group 1, 129 out of 633 (20.38%, 129/633) cases had abnormal CMA findings (Table 2). There were 15 cases (2.37%, 15/633) (Cases 1–15) with clinically significant CNVs (Table 3). Fifteen clinically significant CNVs sized from 0.12 to 5.79 Mb included 11 pathogenic CNVs and 4 likely pathogenic CNVs. Among them, 3 cases were relevant to a known syndrome with variable clinical phenotype: 1q21.1 recurrent microdeletion syndrome (Case 1), 1q21.1 recurrent microduplication syndrome (case 2), and 22q11 duplication syndrome (Case 12). There were 4 cases whose CNVs regions completely covered the known recurrent region (Cases 3, 6, 9 and 10) and 3 cases whose CNVs regions covered or partial covered the known recurrent region (Cases 7, 8 and 14). The ClinGen database suggests that there was some evidence of haploinsufficiency or triplosensitivity for the known related recurrent regions of these 7 cases, and its clinical phenotype is diverse. Case 4 had a 0.12-Mb deletion in the 6q26 region, containing 1 OMIM gene (PARK2). This deletion may be related to autism. Case 5 showed a 0.29-Mb deletion in the 14q23.3 region, which may be related to the risk of neuropsychiatric phenotypes (eg, autism spectrum disorders, seizures, and schizophrenia). Case 11 had a 0.32-Mb deletion in the 21q22.12 region, containing the OMIM gene RUNX1, which is the pathogenic gene of acute myeloid leukemia or familial thrombocytopenia (with myeloid malignancy). Cases 13 and 15 revealed a 1.54-Mb duplication and a 3.02-Mb deletion in Xp22.31 and Xp22.33, respectively, containing 4 and 29 OMIM genes, respectively.
In group 2, the frequency of abnormal CMA findings was 21.07% (158/750) (Table 2), which was similar to group 1 (P>0.05). Fifteen cases (Cases 16–30) with clinically significant CNVs (Table 4) were detected; the detection rate was 2.0% (15/750), and it showed no statistically significant differences from group 1 (2.0% vs 2.37%, P>0.05). Fifteen clinically significant submicroscopic CNVs, sized from 0.19 to 4.70 Mb, included 10 pathogenic CNVs and 5 likely pathogenic CNVs. Six cases were associated with known syndrome with variable penetrance: 1q21.1q21.2 recurrent microdeletion syndrome (Case 16), 1q21.1q21.2 recurrent microduplication syndrome (Cases 17–19), 22q11.21 microdeletion syndrome (Case 28), and 22q11.21 microduplication syndrome (Case 29). Three cases whose CNVs regions covered or completely covered the known recurrent region (Cases 25, 27, and 30). There were 2 other cases whose CNVs regions contained OMIM gene of known clinical syndromes (Cases 20 and 24) and 1 case whose CNVs region covered the most region of Hereditary Liability to Pressure Palsies syndrome (Case 26). Two cases had similar pathogenic CNVs region to those reported in the DECIPHER database (Case s21 and 23). Case 25 revealed a 3.79 Mb deletion in the 5q14.2q14.3 region, containing 5 OMIM genes.
In group 3, there were 43 (31.16%, 43/138) cases had abnormal CMA findings; the detection rate was significantly higher than that in group 1 and group 2. These 43 cases involved 7 cases (5.07%, 7/138) of clinically significant CNVs (Table 5, Cases 31–37), 9 cases (6.52%, 9/138) of benign or likely benign CNVs, and 27 cases (19.57%, 27/138) of VUS. The frequency of clinically significant findings in group 3 was significantly higher than that in group 1 and group 2 (5.07% vs 2.37% and 2.0%, P<0.01). Seven clinically significant submicroscopic CNVs sized from 0.20 to 9.78 Mb included 3 pathogenic CNVs and 4 likely pathogenic CNVs (Table 5). In these 7 cases, 4 cases had similar pathogenic CNVs region to those reported in DECIPHER or ISCA database (Cases 31–33 and 35), 2 cases whose CNVs regions completely or incompletely covered the known recurrent region (Cases 34 and 36), and 1 case revealed a 0.20 Mb duplication in the Xp21.1 region (Case 37), containing the OMIM gene DMD, which is the pathogenic gene of Duchenne muscular dystrophy.
PREGNANCY OUTCOMES:
In 37 cases of clinically significant CNVs, 22 pregnant women (59.46%, 22/37) decided to continue the pregnancy and had successful live births, and 15 pregnant women (40.54%, 15/37) chose to terminate the pregnancy. No abnormal clinical finding was found after the 22 fetuses were born and no neonates had the karyotype reexamined. The results of postnatal follow-up showed that these 22 neonates were born healthy.
Discussion
CMA has been recommended to be performed for prenatal diagnosis of fetuses with ultrasound structural anomalies in China [13]. However, whether to routinely offer CMA to AMA pregnant women or fetuses with USG soft markers when undergoing invasive prenatal diagnosis is uncertain. Moreover, the clinical application of CMA in fetuses with USG soft markers and a normal karyotype has rarely been reported. Hence, the purpose of the present research was to evaluate the application value of CMA in the prenatal diagnosis of AMA pregnant women with or without fetal USG soft markers and isolated fetal USG soft markers when the fetuses had a normal karyotype and to provide further evidence for clinical genetic counseling practice.
The risk of miscarriage caused by amniocentesis is 0.5~1% and it should be routinely explained to all pregnant couples before amniocentesis is performed. On the other hand, the actual rate of miscarriage is very low in our center (<0.1%), so the patients will generally have less fear of miscarriage after preoperative consultation. The results of postoperative follow-up showed that no pregnant women in this study had miscarriage due to amniocentesis. Our study results showed a significant difference in gestational age at amniocentesis between the 3 research groups. The gestational ages at amniocentesis of group 2 and group 3 were significantly higher than in group 1. This may be closely related to the choices of pregnant women and gestational weeks of ultrasonic screening. In general, AMA women can make an appointment for amniocentesis before receiving ultrasound screening, and they preferred amniocenteses because it is more acceptable. Accordingly, AMA women underwent amniocentesis at the correct early time. Pregnant women with fetal USG soft markers, by contrast, can only make an appointment for amniocentesis after ultrasound screening. Moreover, some USG soft markers were only found in ultrasound screening after the 20th gestational week.
In our study, CMA results proved that the detection rate of clinically significant CNVs was 2.37% in group 1, which is higher than the 0.92% reported by Wu et al [14]. Previous research reports showed that the incremental diagnostic rate of CMA in the group of pregnant women with AMA alone ranged from 0.29% to 1.73%, as shown in Table 6. A previous meta-analysis found that the rate of clinically significant CNV was 0.84% (1/119), referred for AMA and parental anxiety [15]. The difference in the detection rate of previous studies may be related to regional or ethnic differences.
Fetal USG soft marker refers to a non-specific marker, which is different from ultrasound structural abnormality and may also be a normal variation. However, previous studies had shown that the fetus with USG soft markers is related to an increased risk of chromosomal abnormalities. Previous studies have shown that the incremental diagnostic rate of CMA for fetuses with USG soft markers and a normal karyotype ranged from 2.60% to 4.95% (Table 6) [7,9,14,16–24]. In this study, the rate was 2.0% and the lower detection rate may be caused by the types of soft markers. A higher detection rate of 7.69% (5/65) for pathogenic CNV has been reported in the fetuses with thickened nuchal fold [22]. Additionally, the clinical application of CMA in fetuses with increased NT and a normal karyotype is a research hotspot. Armour et al recommend that CMA should be offered for fetuses with increased NT (NT ≥3.5 mm) in prenatal diagnosis [10]. Su et al reported that CMA improved the diagnostic rate of chromosomal abnormalities for fetuses with NTs of 2.5–3.4 mm, whose karyotypes were normal [9]. Based on the information presented above, CMA testing should be recommended for fetuses with USG soft markers and a normal karyotype.
Moreover, this study also analyzed the CMA results of the AMA pregnant women with the fetal USG soft markers and a normal karyotype (group 3). There is only 1 previous study that assessed this issue, showing a detection rate of 1.49% (2/134) for abnormal CMA findings with clinical significance [14]. In our study, the detection rate was 5.07% (7/138), which was higher than that reported by Wu et al and similar to that of fetuses with structural anomalies [14].
In 37 cases of clinically significant CNVs, 20 cases (54.05%) were microdeletions, and the remaining 17 cases (45.95%) were microduplications. It is worth noting that 83.78% (31/37) of clinically significant microdeletions or microduplications were associated with mental or/and neurological dysplasia. Recent studies have found that susceptibility locus (SL) for the neurodevelopmental disorder is a fairly common finding in fetuses with submicroscopic CNVs [21]. The SL is also generally associated with autism, developmental delay, intellectual disability, or malformation. In our study, the main SLs were 1q21.1, 15q11.2, 16p13.11, and Xp22.31. These recurrent reciprocal submicroscopic CNVs have been reported to be related to variable clinical phenotype, and some individuals have a normal phenotype, so no clinically recognizable syndrome can be defined [25–27]. This brings some challenges to clinical genetic counseling. In consideration of the association between SL and clinical phenotypes, genetic consultants and pregnant couples should be fully aware of the differences between different abnormal CNVs during clinical genetic counseling. In our research groups, more than half (59.46%, 22/37) of pregnant women whose fetuses had clinically significant CNVs decided to continue the pregnancy and all neonates were born healthy. However, some clinical manifestations of these neonates who were born healthy may be found in the future because we have only followed up 3 months after the birth. Moreover, some clinical manifestations associated with clinically significant CNVs are usually difficult to detect in infancy, such as neurodevelopmental disorder and developmental delay. In addition, we also noted that most AMA pregnant women or fetuses with clinically significant CNVs which are not associated with known syndrome chose to continue pregnancy, while pregnant women aged under 35 years with isolated USG soft markers may be more likely to terminate pregnancy. The results of this research indicate that continued pregnancy may be the most common choice of more pregnant women after they have a clear understanding of the content of genetic counseling.
Nowadays, more and more pregnant women accept and choose the CMA test at their own expense. However, the interpretation of VUS results has become a difficult problem in the clinical application of CMA. The rate of VUS detected by CMA is quite variable [14,28,29]. In our study, VUS was detected from the 3 research groups and had a comparatively higher detection rate of 15.78% than that in the previous literature. The higher detection rate of VUS in our study may be related to the population of pregnant women and the principle of CMA reporting. If submicroscopic anomalies detected by CMA contain a pathogenic gene, according to the relevant regulations of our center, it will be described in the CMA report. From the perspective of the patient, they certainly want to eliminate as many abnormalities as possible and understand the actual results to make sure that the fetus is healthy or normal. At this time, the patient’s mood is complex and contradictory. Therefore, genetic counseling about post-testing is very important. Future research should focus on how to manage the CMA results and how to interpret the abnormal results detected by CMA so that we can accumulate more evidence for genetic counseling and greatly reduce the heavy burdens of genetic consultants or make the patients feel more secure.
Conclusions
In conclusion, in AMA women and fetuses with the USG soft markers, CMA can improve the detection rate of chromosomal aberration by 2.0~5.07% compared with the conventional karyotyping. For this reason, the CMA test should be available to AMA women and fetuses with USG soft markers undergoing prenatal diagnosis. Actually, CMA technology is being promoted in clinical practice, and the limitation of conventional karyotyping is being overcome by such molecular genetic techniques.
Tables
Table 1. Clinical characteristics of the women compared among groups.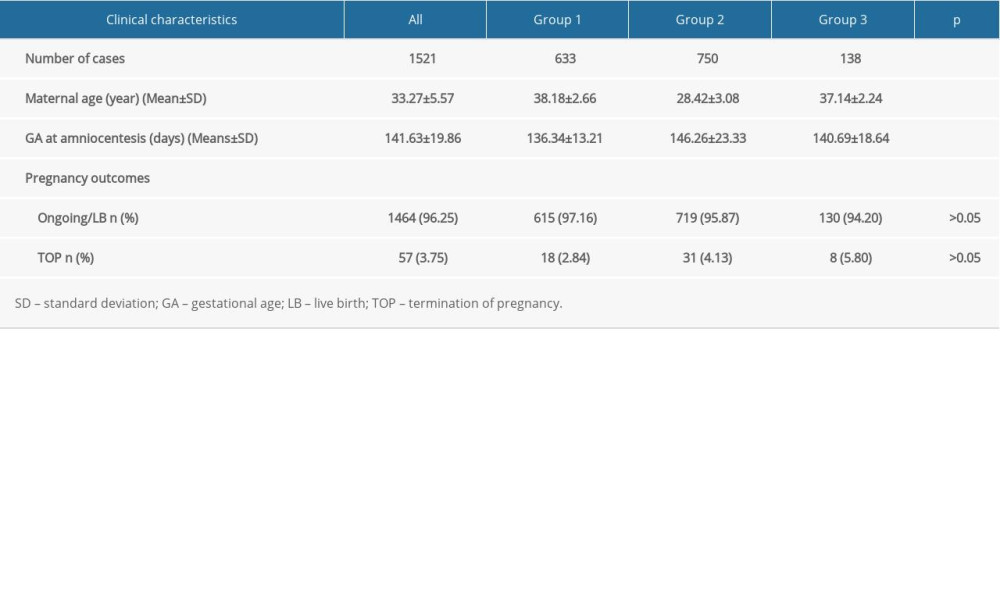 Table 2. Summary of the detection rates of CMA from pregnancies with AMA and fetuses with USG soft markers.
Table 2. Summary of the detection rates of CMA from pregnancies with AMA and fetuses with USG soft markers.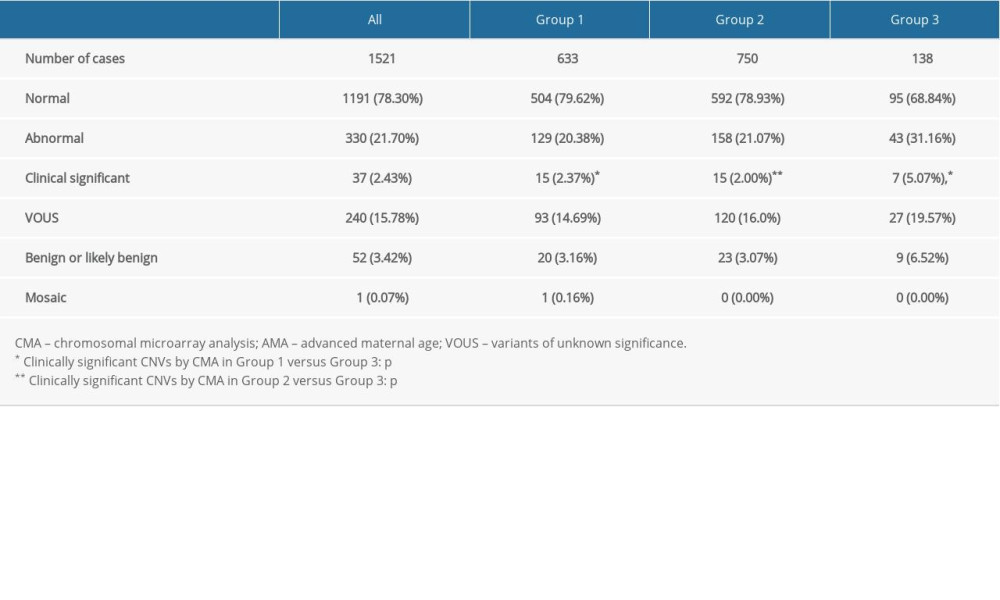 Table 3. List of clinically significant CMA findings for group 1.
Table 3. List of clinically significant CMA findings for group 1.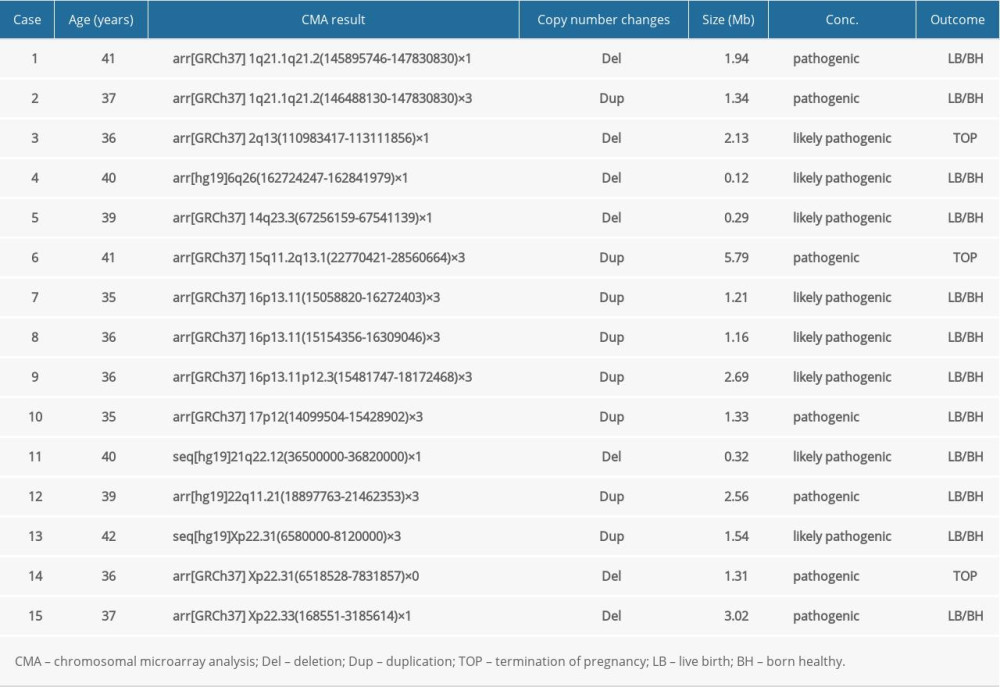 Table 4. List of clinically significant CMA findings for group 2.
Table 4. List of clinically significant CMA findings for group 2.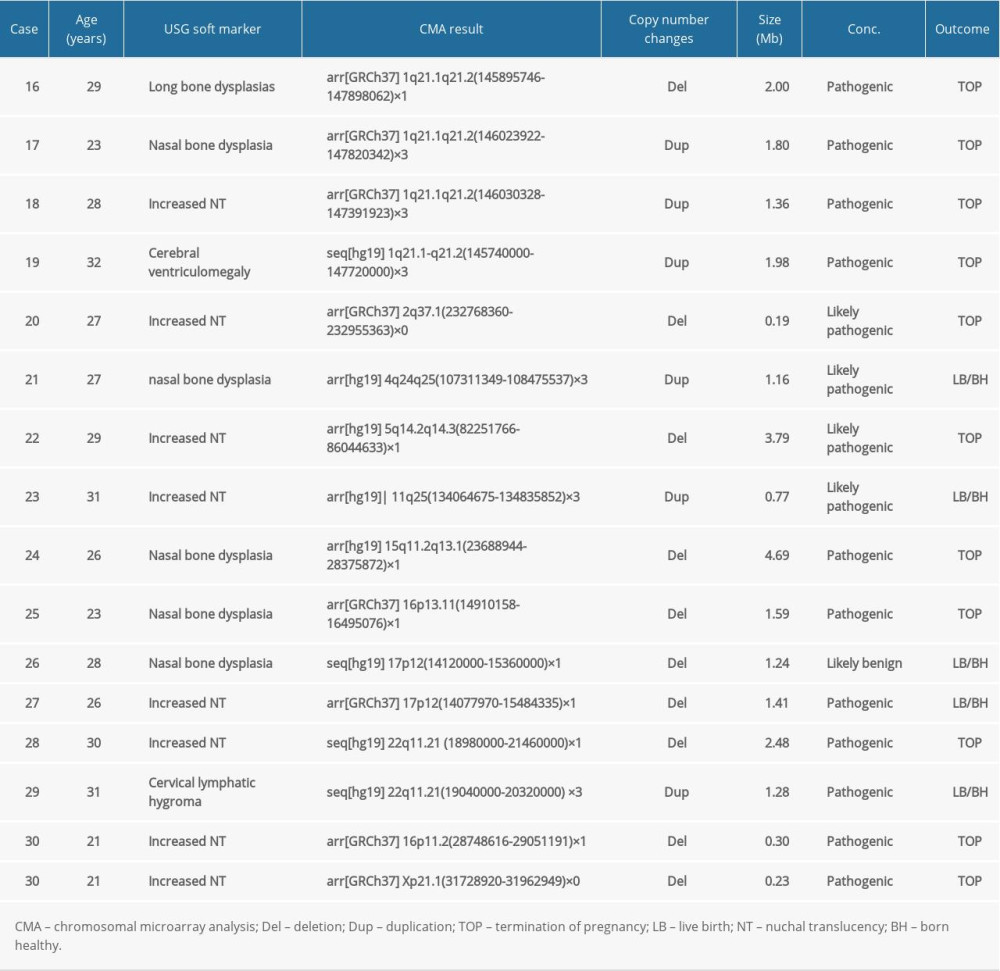 Table 5. List of clinically significant CMA findings for group 3.
Table 5. List of clinically significant CMA findings for group 3.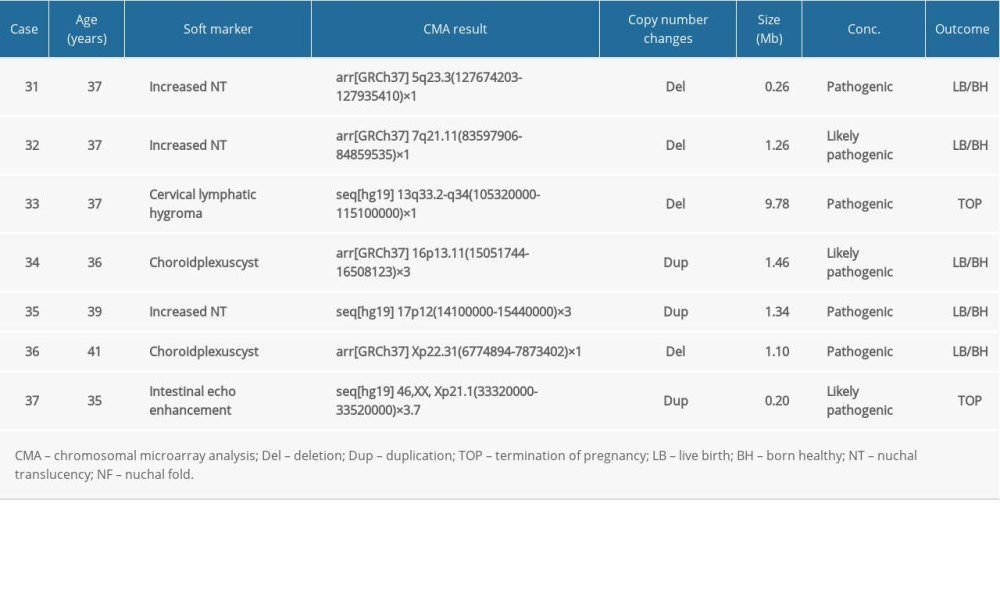 Table 6. Summary of detection rate of clinically significant CMA findings in AMA women and fetal USG soft markers in the literature.
Table 6. Summary of detection rate of clinically significant CMA findings in AMA women and fetal USG soft markers in the literature.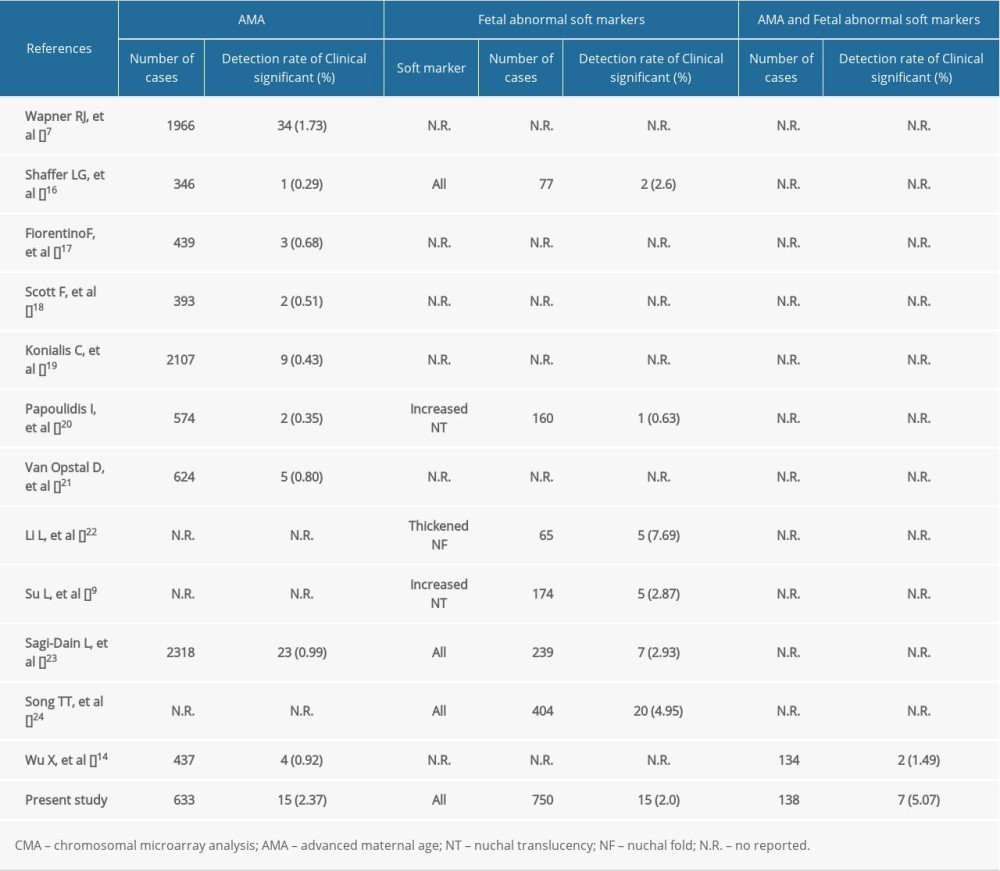
References
1. Kim YJ, Lee JE, Kim SH, Maternal age-specific rates of fetal chromosomal abnormalities in Korean pregnant women of advanced maternal age: Obstet Gynecol Sci, 2013; 56(3); 160-66
2. Wang R, Yu Y, Xi Q, Analysis of prenatal diagnosis before and after implementation of the two-child policy in northeastern China: Medicine (Baltimore), 2019; 98(38); e17200
3. Smeets DF, Historical prospective of human cytogenetics: From microscope to microarray: Clin Biochem, 2004; 37(6); 439-46
4. Reddy UM, Page GP, Saade GR, Karyotype versus microarray testing for genetic abnormalities after stillbirth: N Engl J Med, 2012; 367(23); 2185-93
5. Zhu Y, Lu S, Bian X, A multicenter study of fetal chromosomal abnormalities in Chinese women of advanced maternal age: Taiwan J Obstet Gynecol, 2016; 55(3); 379-84
6. Hillman SC, McMullan DJ, Hall G, Use of prenatal chromosomal microarray: Prospective cohort study and systematic review and meta-analysis: Ultrasound Obstet Gynecol, 2013; 41(6); 610-20
7. Wapner RJ, Martin CL, Levy B, Chromosomal microarray versus karyotyping for prenatal diagnosis: N Engl J Med, 2012; 367(23); 2175-84
8. Ahman A, Axelsson O, Maras G, Ultrasonographic fetal soft markers in a low-risk population: Prevalence, association with trisomies and invasive tests: Acta Obstet Gynecol Scand, 2014; 93(4); 367-73
9. Su L, Huang H, An G, Clinical application of chromosomal microarray analysis in fetuses with increased nuchal translucency and normal karyotype: Mol Genet Genomic Med, 2019; 7(8); e811
10. Armour CM, Dougan SD, Brock JA, Practice guideline: Joint CCMG-SOGC recommendations for the use of chromosomal microarray analysis for prenatal diagnosis and assessment of fetal loss in Canada: J Med Genet, 2018; 55(4); 215-21
11. Shaffer L, Slovak M, Campbell L: ISCN 2013 : An international system for human cytogenetic nomenclature, 2013; 138, Basel, Switzerland, S Karger
12. South ST, Lee C, Lamb ANWorking Group for the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee, ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: Revision 2013: Genet Med, 2013; 15(11); 901-9
13. Qingwei Qi HWExpert consensus on the application of chromosome microarray analysis in prenatal diagnosis: Chinese Journal of Obstetrics and Gynecology, 2014; 049(8); 570-72 [in Chinese]
14. Wu X, An G, Xie X, Chromosomal microarray analysis for pregnancies with or without ultrasound abnormalities in women of advanced maternal age: J Clin Lab Anal, 2020; 34(4); e23117
15. Srebniak MI, Joosten M, Knapen M, Frequency of submicroscopic chromosomal aberrations in pregnancies without increased risk for structural chromosomal aberrations: Systematic review and meta-analysis: Ultrasound Obstet Gynecol, 2018; 51(4); 445-52
16. Shaffer LG, Dabell MP, Fisher AJ, Experience with microarray-based comparative genomic hybridization for prenatal diagnosis in over 5000 pregnancies: Prenat Diagn, 2012; 32(10); 976-85
17. Fiorentino F, Caiazzo F, Napolitano S, Introducing array comparative genomic hybridization into routine prenatal diagnosis practice: A prospective study on over 1000 consecutive clinical cases: Prenat Diagn, 2011; 31(13); 1270-82
18. Scott F, Murphy K, Carey L, Prenatal diagnosis using combined quantitative fluorescent polymerase chain reaction and array comparative genomic hybridization analysis as a first-line test: Results from over 1000 consecutive cases: Ultrasound Obstet Gynecol, 2013; 41(5); 500-7
19. Konialis C, Pangalos C, Dilemmas in prenatal chromosomal diagnosis revealed through a single center’s 30 years’ experience and 90,000 cases: Fetal Diagn Ther, 2015; 38(3); 218-32
20. Papoulidis I, Sotiriadis A, Siomou E, Routine use of array comparative genomic Hybridization (aCGH) as standard approach for prenatal diagnosis of chromosomal abnormalities. Clinical experience of 1763 prenatal cases: Prenat Diagn, 2015; 35(13); 1269-77
21. Van Opstal D, de Vries F, Govaerts L, Benefits and burdens of using a SNP array in pregnancies at increased risk for the common aneuploidies: Hum Mutat, 2015; 36(3); 319-26
22. Li L, Fu F, Li R, Prenatal diagnosis and pregnancy outcome analysis of thickened nuchal fold in the second trimester: Medicine (Baltimore), 2018; 97(46); e13334
23. Sagi-Dain L, Cohen Vig L, Kahana S, Chromosomal microarray vs. NIPS: Analysis of 5541 low-risk pregnancies: Genet Med, 2019; 21(11); 2462-67
24. Song T, Zhang J, Dang Y, Application value of chromosomal microarray analysis in prenatal diagnosis of fetuses with ultrasonographic soft markers: Medical Journal of West China, 2019; 31(4); 513-17
25. Nagai K, Shima H, Kamimura M, Xp22.31 microdeletion due to microhomology-mediated break-induced replication in a boy with contiguous gene deletion syndrome: Cytogenet Genome Res, 2017; 151(1); 1-4
26. de Kovel CG, Trucks H, Helbig I, Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies: Brain, 2010; 133(Pt 1); 23-32
27. Buse M, Cuttaia HC, Palazzo D, Expanding the phenotype of reciprocal 1q21.1 deletions and duplications: A case series: Ital J Pediatr, 2017; 43(1); 61
28. Oneda B, Baldinger R, Reissmann R, High-resolution chromosomal microarrays in prenatal diagnosis significantly increase diagnostic power: Prenat Diagn, 2014; 34(6); 525-33
29. Ganesamoorthy D, Bruno DL, McGillivray G, Meeting the challenge of interpreting high-resolution single nucleotide polymorphism array data in prenatal diagnosis: Does increased diagnostic power outweigh the dilemma of rare variants?: BJOG, 2013; 120(5); 594-606
Tables
 Table 1. Clinical characteristics of the women compared among groups.
Table 1. Clinical characteristics of the women compared among groups. Table 2. Summary of the detection rates of CMA from pregnancies with AMA and fetuses with USG soft markers.
Table 2. Summary of the detection rates of CMA from pregnancies with AMA and fetuses with USG soft markers. Table 3. List of clinically significant CMA findings for group 1.
Table 3. List of clinically significant CMA findings for group 1. Table 4. List of clinically significant CMA findings for group 2.
Table 4. List of clinically significant CMA findings for group 2. Table 5. List of clinically significant CMA findings for group 3.
Table 5. List of clinically significant CMA findings for group 3. Table 6. Summary of detection rate of clinically significant CMA findings in AMA women and fetal USG soft markers in the literature.
Table 6. Summary of detection rate of clinically significant CMA findings in AMA women and fetal USG soft markers in the literature. In Press
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








