26 November 2020: Database Analysis
The Clinical Significance of Dickkopf Wnt Signaling Pathway Inhibitor Gene Family in Head and Neck Squamous Cell Carcinoma
Yuan Hu1ABCEF, Muyuan Liu1CD, Shaowei Xu1BD, Shaokun Li1EF, Mingfeng Yang1G, Tian Su1B, Zihong Yuan1C, Hanwei Peng1AD*DOI: 10.12659/MSM.927368
Med Sci Monit 2020; 26:e927368
Abstract
BACKGROUND: Dickkopf Wnt signaling pathway inhibitor (DKK) gene family, which is known to inhibit the Wnt regulation process, is widely found in cancers. However, the roles and functions of specific family members in head and neck squamous cell carcinoma (HNSCC) are still unclear.
MATERIAL AND METHODS: Online bioinformatics tools (Oncomine, UALCAN, Kaplan-Meier plotter, GEPIA, Metascape, and STRING) were used to analyze the relationships between distinct DKKs and HNSCC. The transcriptome expression, clinical association, functions, pathways, and protein-protein interaction networks of DKKs in HNSCC were explored.
RESULTS: The mRNA expression of DKK1, DKK3, and Dickkopf-like acrosomal protein 1 (DKKL1) in HNSCC was significantly higher than in normal tissues, while that of DKK4 was lower. The mRNA expression of DKK1, DKK3, and DKKL1 was elevated in higher-grade HNSCC. The mRNA expression of DKK1 and DKK3 was elevated in human papillomavirus (HPV)-negative HNSCC, while DKKL1 had a higher mRNA expression in HPV-positive HNSCC. In addition, DKK1 was significantly associated with unfavorable overall survival in HNSCC patients. DKK3 was more likely to be a negative factor for the 5-year survival rate, while DKK4 was the opposite. DKK1 function was mainly enriched in GTPase-mediated signal transduction. Porcupine O-acyltransferase, a key regulator of the Wnt signaling pathway, was also associated with DKK1 in the protein-protein interaction network.
CONCLUSIONS: With regard to improving the therapeutic strategies of HNSCC in the future, DKK1 could be an unfavorable prognostic biomarker. DKK3, DKK4, and DKKL1 might be potential biomarkers for HNSCC.
Keywords: Carcinoma, Squamous Cell, Gene Expression Profiling, Head and Neck Neoplasms, Adaptor Proteins, Signal Transducing, Intercellular Signaling Peptides and Proteins, Multigene Family, Protein Interaction Maps, RNA, Messenger, Squamous Cell Carcinoma of Head and Neck
Background
Head and neck squamous cell carcinoma (HNSCC) has been reported to be the sixth most common malignancy worldwide. The most recent cancer statistics estimated that approximately 65 410 new cases and 14 620 deaths due to HNSCC were projected to occur in the United States in 2019 [1]. Chronic exposure to tobacco, tobacco-like products, and alcohol are well known to increase the risk of HNSCC [2]. Moreover, due to the emergence of a distinct subset of HPV-positive oropharyngeal cancers, the epidemiology of HNSCC has changed dramatically in the past 2 decades [3].
In the past 30 years, the overall survival (OS) of HNSCC patients has not changed significantly. The 5-year OS of patients with locally advanced disease is only 50%, and the OS of patients with relapsed or metastatic disease is approximately 6 months [4]. Therefore, an urgent need exists to uncover the molecular mechanisms of HNSCC and develop new diagnostic and/or therapeutic targets to facilitate early detection and progression monitoring and improve treatment outcomes.
The Dickkopf Wnt signaling pathway inhibitor (DKK) gene family, which comprises 4 members (DKK1-4) and a specific DKK3-related gene (Dickkopf-like acrosomal protein 1 [DKKL1]), is related to cancers through inhibition of the Wnt regulation process [5]. DKK1 has been reported to be increased in patient serum, and overexpression of DKK1 was associated with a decrease in the OS in patients with pancreatic ductal adenocarcinoma [6]. In HNSCC, patients with DKK1 mutations had a lower distant metastasis rate and a longer disease-free survival rate [7]. Meanwhile, DKK3 was found to play a significant role in tumor suppression in pancreatic cancer, prostate cancer, gastric cancer, head and neck cancer (HNC), breast cancer, and testicular cancer [8].
However, the roles of specific DKK family members in the development and progression of HNSCC have not yet been fully elucidated. We analyzed the expression of different DKK family members in patients with HNSCC and the relationships between their expression and clinical parameters, using the online bioinformatics tools Oncomine, UALCAN, Kaplan-Meier plotter, GEPIA, Metascape, and STRING. In addition, we predicted the functions, signaling pathways, and PPI networks of DKKs.
Material and Methods
ETHICS STATEMENT:
Our research protocol was approved by the Ethics Committee of the Cancer Hospital of Shantou University Medical College. All data were obtained from online databases. Additional written informed consent was not required.
ONCOMINE:
Oncomine (https://www.oncomine.org) contains 18,000 cancer gene expression profiles that have been standardized and analyzed to provide high-quality cancer transcriptome data to the biomedical research community [9]. In our study, we utilized the Oncomine database to analyze transcriptional levels of 5 different DKK members between different cancer tissues and their normal control tissues. The statistical differences were investigated by t test (P=0.01; fold change, 1.5; gene rank, 10%; data type, mRNA).
UALCAN:
UALCAN (http://ualcan.path.uab.edu) is an interactive website that utilizes TCGA level-3 RNA-sequencing and clinical data from 31 cancer types to conduct in-depth analyses of TCGA gene expression data [10]. In this study, we used UALCAN to study the mRNA expression of 5 DKK family members in primary HNSCC tissues and their relationships with clinicopathological parameters. The differences in transcriptional level were compared by t test, and P<0.05 was considered statistically significant.
KAPLAN-MEIER PLOTTER:
The prognostic value of the mRNA expression of distinct DKKs in HNSCC was investigated through the Kaplan-Meier plotter (http://kmplot.com/analysis/) [11]. In the Kaplan-Meier plotter, cancer patients are divided into high- and low-expression groups based on the median values for mRNA expression, and the groups are compared on a Kaplan-Meier survival curve. The differences are assessed by log-rank testing, and they are considered statistically significant if P<0.05.
GEPIA:
GEPIA (http://gepia.cancer-pku.cn/) is a new web-based tool that provides fast and customizable functions based on TCGA and genotype-tissue expression data [12]. In this study, GEPIA was used to detect the top 20 genes similar to 5 DKK family members in HNSCC for further exploration.
METASCAPE:
Metascape (https://metascape.org) is an interactive portal designed to provide comprehensive gene list annotation and analysis resources for research [13]. In this study, Metascape was used to perform Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis for each DKK gene and the similar genes detected by GEPIA. The P value was set to 0.05, and the minimum enrichment value was set to 3.
STRING:
STRING (https://string-db.org/) aims to collect, score, and integrate all publicly accessible protein-protein interaction (PPI) data sources, and it predicts their potential functions [14]. In this study, we performed a PPI network analysis of all DKKs and similar genes to explore their interactions.
Results
DIFFERENTIAL EXPRESSION OF 5 DKK FAMILY MEMBERS IN PATIENTS WITH HNSCC:
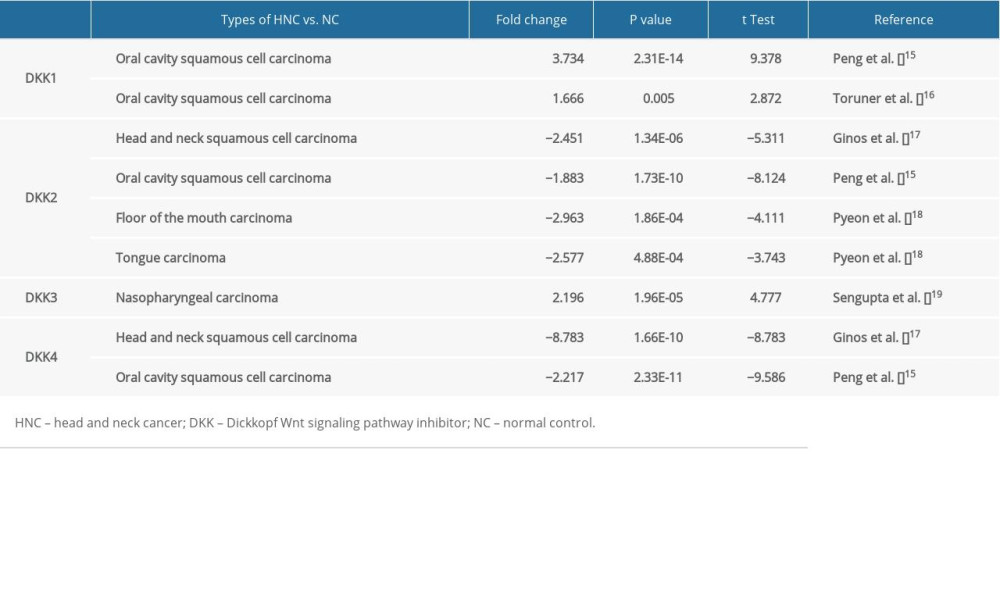
First, the mRNA expression level was studied through the Oncomine and UALCAN. As shown in Figure 1 and Table 1, the mRNA expression levels of 5 DKK family members were measured and compared with normal tissues for different kinds of cancers, using the Oncomine database. Compared with the normal controls, the mRNA expression of DKK1 and DKK3 in HNC was significantly higher, while the mRNA expression of DKK2 and DKK4 was significantly lower. In HNSCC, DKK1 was overexpressed compared with normal tissues, with a fold change of 3.734 (P=2.31E-14) in the head-neck dataset of Peng et al. [15], while Toruner et al. [16] observed a 1.666-fold increase in mRNA expression of DKK1 in HNSCC samples (P=0.005). For DKK2, significant downregulation was found in HNSCC tissues compared with normal controls. The results from the Ginos et al. [17] dataset showed that there was a −2.451-fold (P=1.34E-06) change in mRNA expression in HNSCC tissues. And, in the Peng et al. [17] dataset, a −1.883-fold change in DKK2 mRNA expression was found in HNSCC tissues compared with normal tissues (P=1.73E-10). Similarly, Ginos et al. [17] found a −8.783-fold change in DKK4 mRNA expression (=1.66E-10), while Peng et al. [15] found a −2.217-fold change for it.
Next, we utilized UALCAN to study the mRNA expression of 5 DKK family members based on the TCGA dataset. As shown in Figure 2, the mRNA for DKK1, DKK3, and DKKL1 (Figure 2A, 2C, 2E) was significantly overexpressed in HNSCC compared with normal tissue, while the DKK4 mRNA expression (Figure 2D) was significantly lower in HNSCC. However, the mRNA expression of DKK2 (Figure 2B) had no statistical difference in the TCGA dataset. After comparing all results from these 2 databases, we found convincing evidence that DKK1 and DKK3 were overexpressed in HNSCC patients at the mRNA level, while DKK4 had low expression.
MRNA EXPRESSION OF DKKS AND CLINICOPATHOLOGICAL PARAMETERS OF HNSCC:
We also analyzed the relationship between the mRNA expression of specific DKK family members and the clinicopathological parameters of HNSCC, including cancer stage, tumor grade, and HPV status, through UALCAN. As shown in Figure 3, the mRNA expression of DKK1, DKK3, DKK4, and DKKL1 (Figure 3A, 3C–3E) did not correlate with tumor stage. For DKK1 (Figure 3A), mRNA expression in stage 1 was significantly associated with the expression in stages 2–4.
However, as shown in Figure 4, the mRNA expression of DKK1, DKK3, and DKKL1 (Figure 4A, 4C–4E) was significantly correlated with tumor grade, and the mRNA expression level of DKKs tended to increase with the tumor grade. The mRNA expression in grade 4 was treated as an unreliable value due to the small sample size (n=7). The expression of DKKL1 had significant correlations with tumor grade, while the expression of DKK2 and DKK4 (Figure 4B, 4D) had no statistical correlation with tumor grade.
The mRNA expression of DKK1 and DKK3 (Figure 5A, 5C) was significantly associated with HPV status, and the level of mRNA expression of DKK1 and DKK3 in HPV-negative patients tended to be higher. On the other hand, the level of DKKL1 mRNA expression was higher in HPV-positive patients (Figure 5E). The mRNA expression of DKK4 (Figure 5D) was not significantly different between HPV-positive and HPV-negative patients. The mRNA expression of DKK2 (Figure 5B) had no statistical correlation with HPV status.
PROGNOSTIC VALUE OF MRNA EXPRESSION OF DKKS IN HNSCC:
We used the Kaplan-Meier plotter to analyze the prognostic value of mRNA expression of distinct DKKs in HNSCC patients. As shown in Figure 6, the mRNA expression of DKK1, DKK3, and DKK4 (Figure 6A, 6C, 6D) was statistically associated with prognosis in HNSCC patients. The mRNA expression of DKK3 and DKK4 (hazard ratio [HR]=1.46, 95% confidence interval [CI]: 1.11–1.92, P=0.0066, and HR=0.76, 95% CI: 0.58–0.99, P=0.045, respectively) had statistical differences for OS of HNSCC patients. However, due to the influence of confounding factors, the survival curves of DKK3 and DKK4 intersected. DKK3 was more likely to be an unfavorable factor in prognosis, while DKK4 was more likely to be a favorable prognostic factor for 5-year survival rate. High expression of DKK1 (HR=2.12, 95% CI: 1.61–2.79, P=4.4e-08) was significantly associated with poor prognosis in HNSCC patients.
FUNCTIONS AND PATHWAYS OF DKKS AND SIMILAR GENES IN HNSCC:
Similar genes may have similar functions. Therefore, we used GEPIA to predict the top 20 genes that were similar to 5 DKK family members in HNSCC. Then, we used Metascape to perform GO enrichment analysis and KEGG pathway enrichment analysis on these genes to predict the functions and pathways of distinct DKKs. As shown in Figure 7, DKK1 function appeared to be mainly related to small GTPase-mediated signal transduction, transmembrane receptor protein tyrosine kinase signaling pathway, and positive regulation of cell death (Figure 7A). DKK2 function appeared to be related to blood vessel development, multicellular organismal homeostasis, and endothelium development (Figure 7B). Cell adhesion molecule binding, extracellular matrix organization, and endoplasmic reticulum lumen could be associated with DKK3 (Figure 7C) function. DKK4 function seemed to be related to receptor regulator activity, receptor ligand activity, and cellular response to hormone stimulus (Figure 7D). A search of GEPIA for genes similar to DKKL1 had no result due to the small gene list (Figure 7E). Due to limited enrichment conditions, there were no outcomes for the KEGG pathway.
PPI NETWORK ANALYSIS OF ALL DKKS AND SIMILAR GENES:
Lastly, to further explore the interactions between DKKs and similar genes, we conducted a PPI network analysis through STRING. Unqualified genes were removed. As shown in Figure 8, DKK1 and DKK2 were associated with every other DKK. DKK1 was also related to parathyroid hormone, fibroblast growth factor, and porcupine O-acyltransferase (PORCN). DKK4 and DKKL1 were related to every other DKK except DKK3. DKK3 was only related to DKK1 and DKK2. Finally, DKKL1 was also related to fibulin 5 (FBLN5).
Discussion
DKKs are associated with many cancers, including pancreatic cancer, prostate cancer, stomach cancer, HNC, breast cancer, and testicular cancer [5–8]. However, the role of different DKK family members in the occurrence and development of HNC remains unknown. To clarify this relationship, we analyzed the expression, clinicopathological relationship, prognostic value, and functions of each DKK family member.
DKK1 is a secreted protein that can inhibit the Wnt signal transduction pathway [21]. Previous studies showed that DKK1 had a proapoptotic effect on tumor cells by inhibiting the classic pathway by downregulating β-catenin [22]. In thyroid cancer, DKK1 inhibited tumor cell survival and migration by regulating Wnt/β-catenin signaling and E-cadherin expression [23]. DKK1 was also found to be overexpressed in hepatocellular carcinoma, and ectopic expression of DKK1 promoted tumor cell migration and invasion at least partly through the β-catenin/MMP7 signal axis [24]. The frequent allele loss of the DKK-1 locus (10q11.2) was reported to be associated with low distant metastasis of HNSCC and better prognosis [7]. In addition, increased DKK1 expression was an independent poor prognostic indicator of survival in HNSCC [25]. In our study, DKK1 was overexpressed in HNSCC compared with normal tissues in the Oncomine and TCGA databases. In addition, the mRNA expression of DKK1 was correlated with tumor grade and HPV status. The mRNA expression level of DKK1 tended to be higher as the tumor grade increased. The mRNA expression of DKK1 in HPV-negative patients also tended to be higher. Higher expression of DKK1 in patients with HNSCC was significantly associated with poor OS. The function of DKK1 was mainly enriched in GTPase-mediated signal transduction, which might indicate the signaling mechanism of DKK1 in HNSCC. PORCN, a key regulator of the Wnt signaling pathway, was also associated with DKK1 in the PPI network. These results are consistent with previous findings in HNSCC and suggest that DKK1 may be an unfavorable prognostic biomarker for HNSCC, thus deserving further study.
To date, little is known about the expression and role of DKK2 in cancers (especially in HNSCC). MicroRNA (miR)-21 was found to be overexpressed and correlated with tumor invasion in tongue cancer, and miR-21 was reported to promote cancer cell invasion via the Wnt/β-catenin pathway by targeting DKK2
Although DKK3 belongs to the DKK family, DKK3 did not seem to antagonize Wnt signaling because DKK3 did not bind to lipoprotein receptor-related or Kremen protein [5], making the biological role of DKK3 unclear. The previous study demonstrated that DKK3 protein expression was widely expressed in HNSCC and its precursor lesions [5]. In addition, patients with DKK3 expression displayed lower disease-free and metastasis-free survival [28]. Knockdown of DKK3 reduced cancer cell migration and invasion independently of the Wnt pathways in oral squamous cell carcinoma-derived cells [29]. But in papillary thyroid carcinoma, knockdown demonstrated that the DKK3 gene was a potential tumor suppressor gene, and aberrant promoter methylation was a significant mechanism for its downregulation [30]. In our study, DKK3 mRNA was overexpressed in the Oncomine and TCGA databases, and it was correlated with tumor grade and HPV status. The higher mRNA expression of DKK3 correlated with an unfavorable prognosis for the 5-year survival rate in HNSCC patients. Of note, the function of DKK3 was mainly enriched in cell adhesion molecule binding, which might reveal its functional mechanism. DKK3 was only related with DKK1 and DKK2 in the PPI network, which suggests that it might not be significantly associated with Wnt pathways. These results demonstrated that DKK3 may participate in cell adhesion molecule binding to regulate HNSCC progression and be a potentially unfavorable prognostic factor for HNSCC.
The impact of DKK4 on the tumor process of HNSCC remains unknown, and few studies have investigated DKK4 in HNSCC. However, DKK4 transcription and translation have been detected in hepatocellular carcinoma, colorectal cancer, gastric cancer, pancreatic cancer, renal cell cancer, ovarian cancer, and esophageal cancer. DKK4 tended to be downregulated in hepatocellular carcinoma, but it was overexpressed in other cancers [31]. In our study, the mRNA expression of DKK4 in HNSCC was significantly downregulated in the Oncomine and TCGA databases. Higher mRNA expression of DKK4 had a favorable prognosis for the 5-year survival rate in HNSCC patients. The function of DKK4 was mostly enriched in receptor regulator activity. These results indicate that DKK4 may be a potential protective factor in HNSCC.
Among all DKKs, the least was known about DKKL1 and its association with cancer. DKKL1 was found to be significantly overexpressed in colorectal cancer compared with healthy colon samples isolated from the same patient, and according to the Dukes staging system, DKKL1 expression was positively associated with disease progression [32]. In our study, no results were found in the Oncomine database, but in the TCGA database, the mRNA expression level of DKKL1 in HNSCC was significantly higher than in normal tissues. The mRNA expression of DKKL1 was also correlated with tumor grade and HPV status. Therefore, the function of DKKL1 in HNSCC remains a mystery. Whether DKKL1 could be a biomarker for HNSCC requires further studies.
There are limitations to our research. First, this was a retrospective study, and all data were retrieved from online databases, which may have led to a decline in reliability and validity. Large-scale prospective research should be conducted in the future to validate our findings. Secondly, our research lacked
Conclusions
In conclusion, this study provided a basic understanding of the expression and clinical value of differently expressed DKKs in HNSCC. Although DKKs belong to the same gene family, their expression levels in HNSCC varied, indicating that they each might have different functions in tumor progression. DKK1, DKK3, and DKK4 were potential biomarkers that warrant further study. We also found that DKK1 was significantly associated with poor OS in HNSCC patients. In addition, we found that DKK3 was more likely to be a negative factor for the 5-year survival rate, while DKK4 was the opposite. We also revealed that the mRNA expression of DKKL1 was overexpressed in HNSCC and significantly related to tumor grade and HPV status. These results provide direction for developing new biomarkers to promote future therapeutic strategies in HNSCC.
Figures
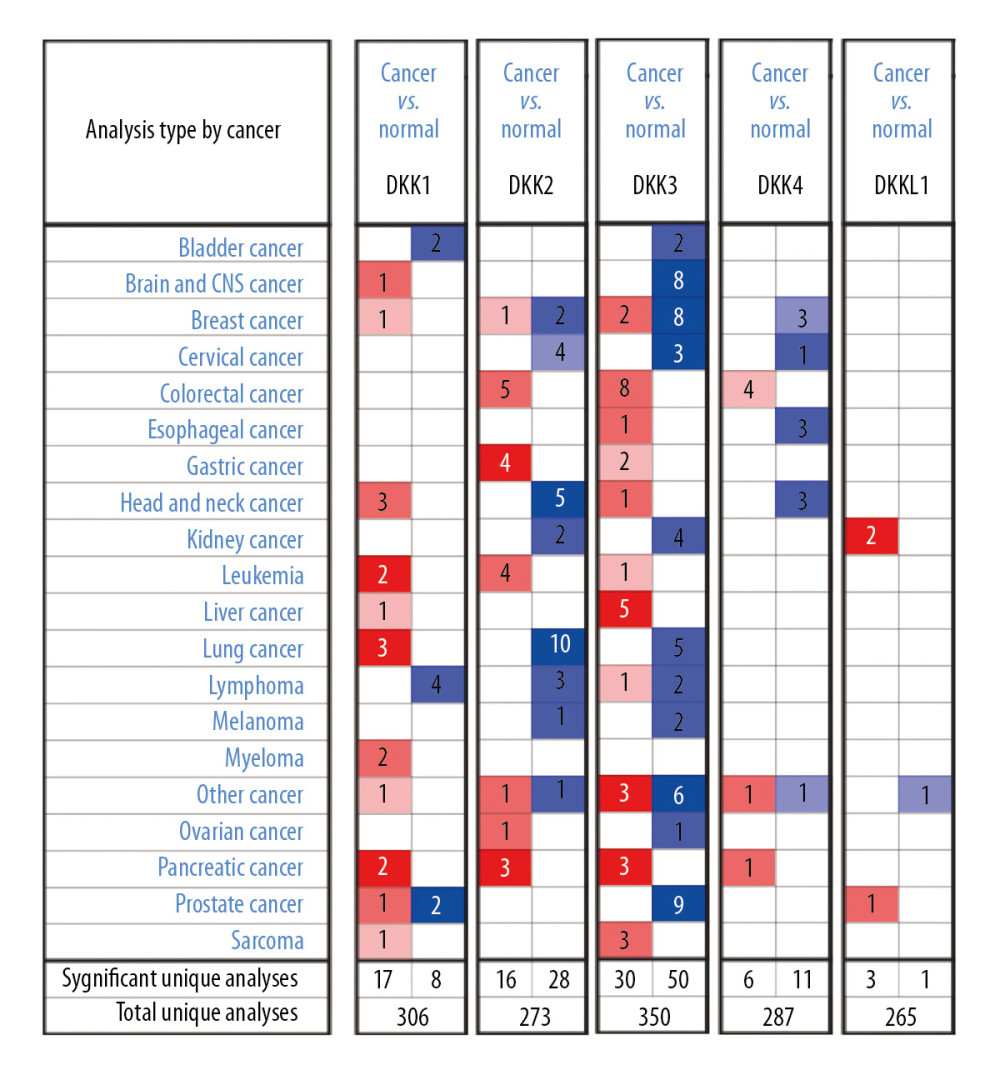 Figure 1. The mRNA expression of DKKs in 20 different types of cancers (Oncomine database). The figure showed the numbers of datasets with statistically significant mRNA overexpression (red) or downregulated expression (blue) of DKKs. The difference in transcriptional expression was analyzed by t test. Cut-off of P value and fold change were as follows: P=0.01; fold change, 1.5; gene rank, 10%; data type, mRNA. DKK – Dickkopf Wnt signaling pathway inhibitor.
Figure 1. The mRNA expression of DKKs in 20 different types of cancers (Oncomine database). The figure showed the numbers of datasets with statistically significant mRNA overexpression (red) or downregulated expression (blue) of DKKs. The difference in transcriptional expression was analyzed by t test. Cut-off of P value and fold change were as follows: P=0.01; fold change, 1.5; gene rank, 10%; data type, mRNA. DKK – Dickkopf Wnt signaling pathway inhibitor. 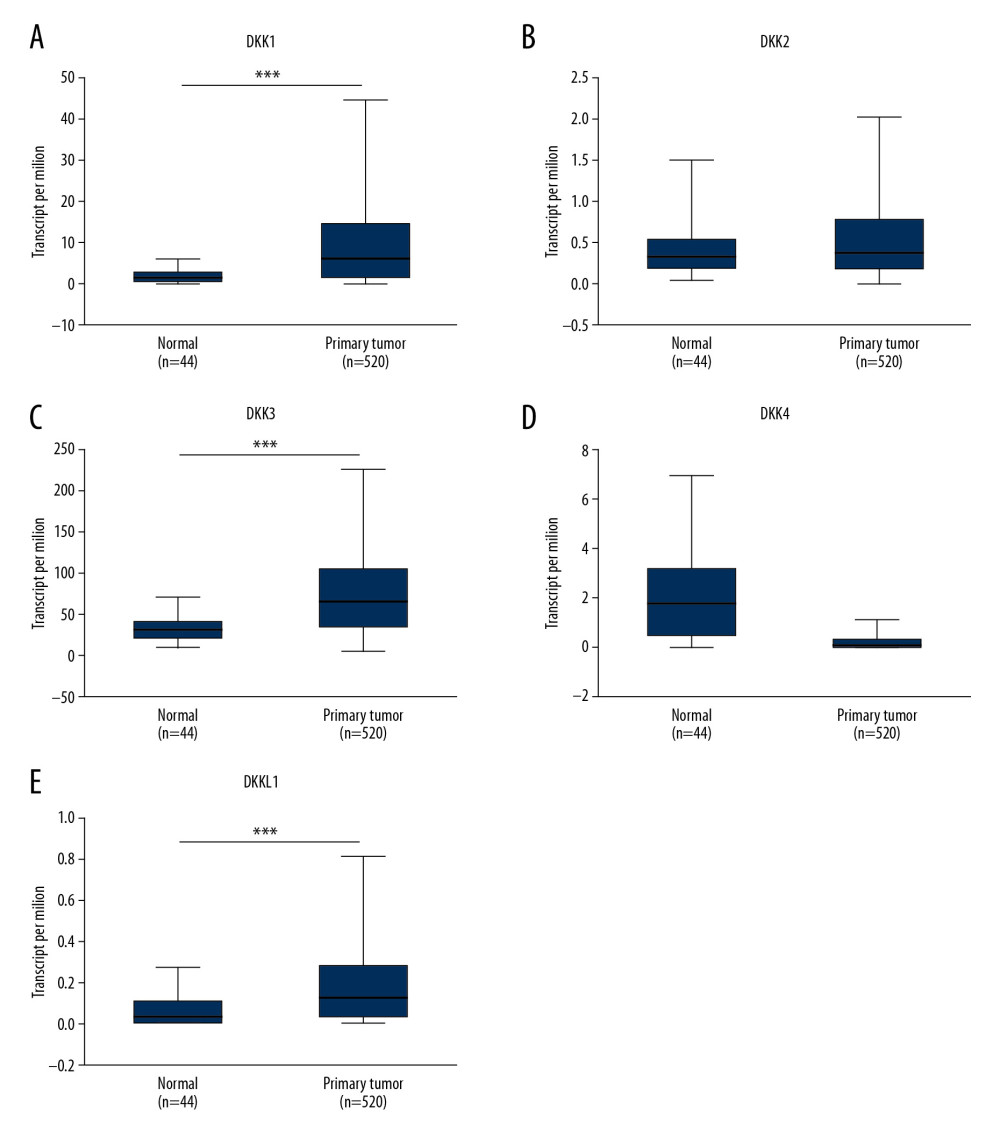 Figure 2. (A–E) The mRNA expressions of distinct DKKs in HNSCC tissues and normal controls (UALCAN). *** P<0.001. DKK, Dickkopf Wnt signaling pathway inhibitor; HNSCC, head and neck squamous cell carcinoma.
Figure 2. (A–E) The mRNA expressions of distinct DKKs in HNSCC tissues and normal controls (UALCAN). *** P<0.001. DKK, Dickkopf Wnt signaling pathway inhibitor; HNSCC, head and neck squamous cell carcinoma. 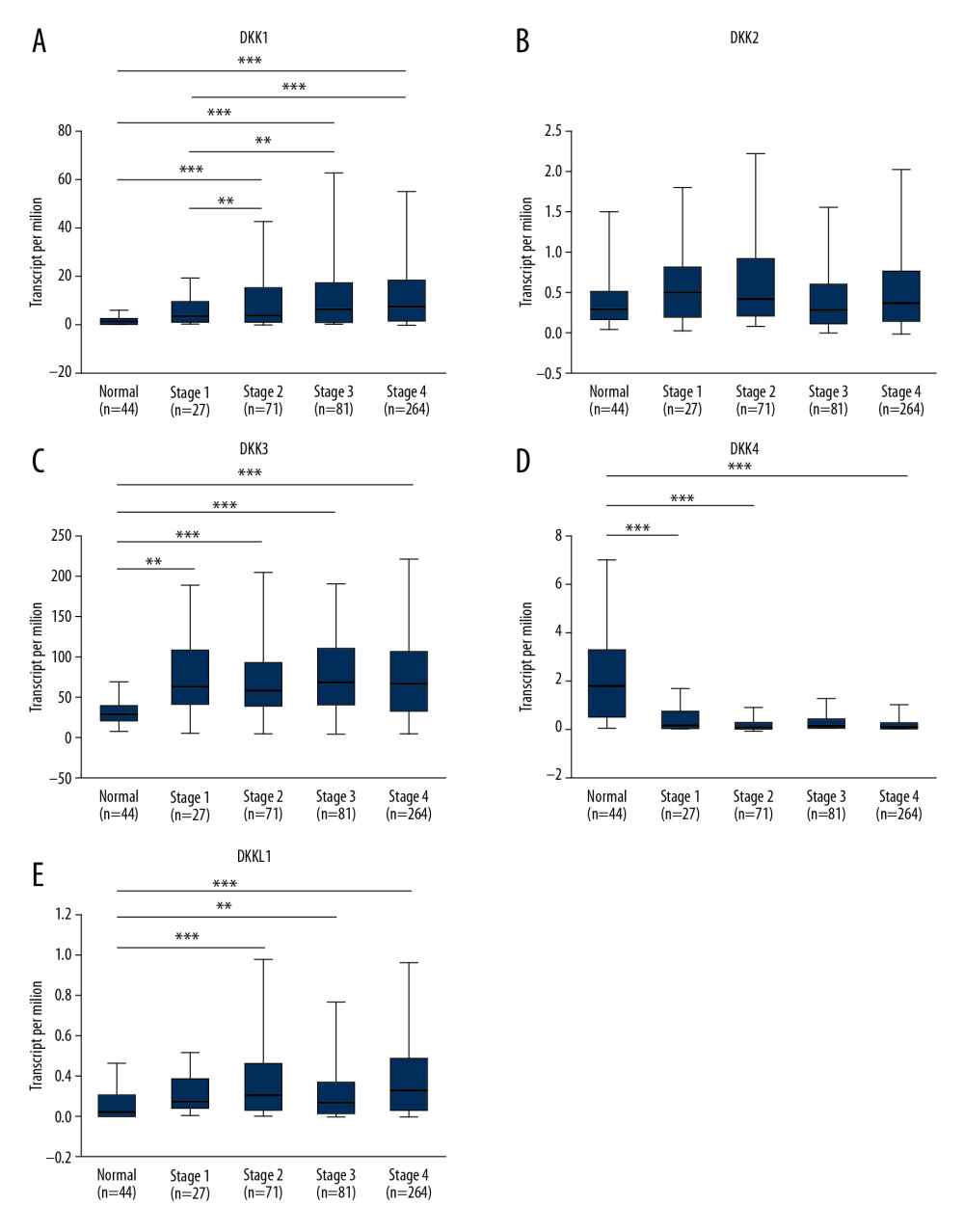 Figure 3. (A–E) Relationship between the mRNA expression of distinct DKKs and tumor stage of HNSCC patients. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 3. (A–E) Relationship between the mRNA expression of distinct DKKs and tumor stage of HNSCC patients. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. 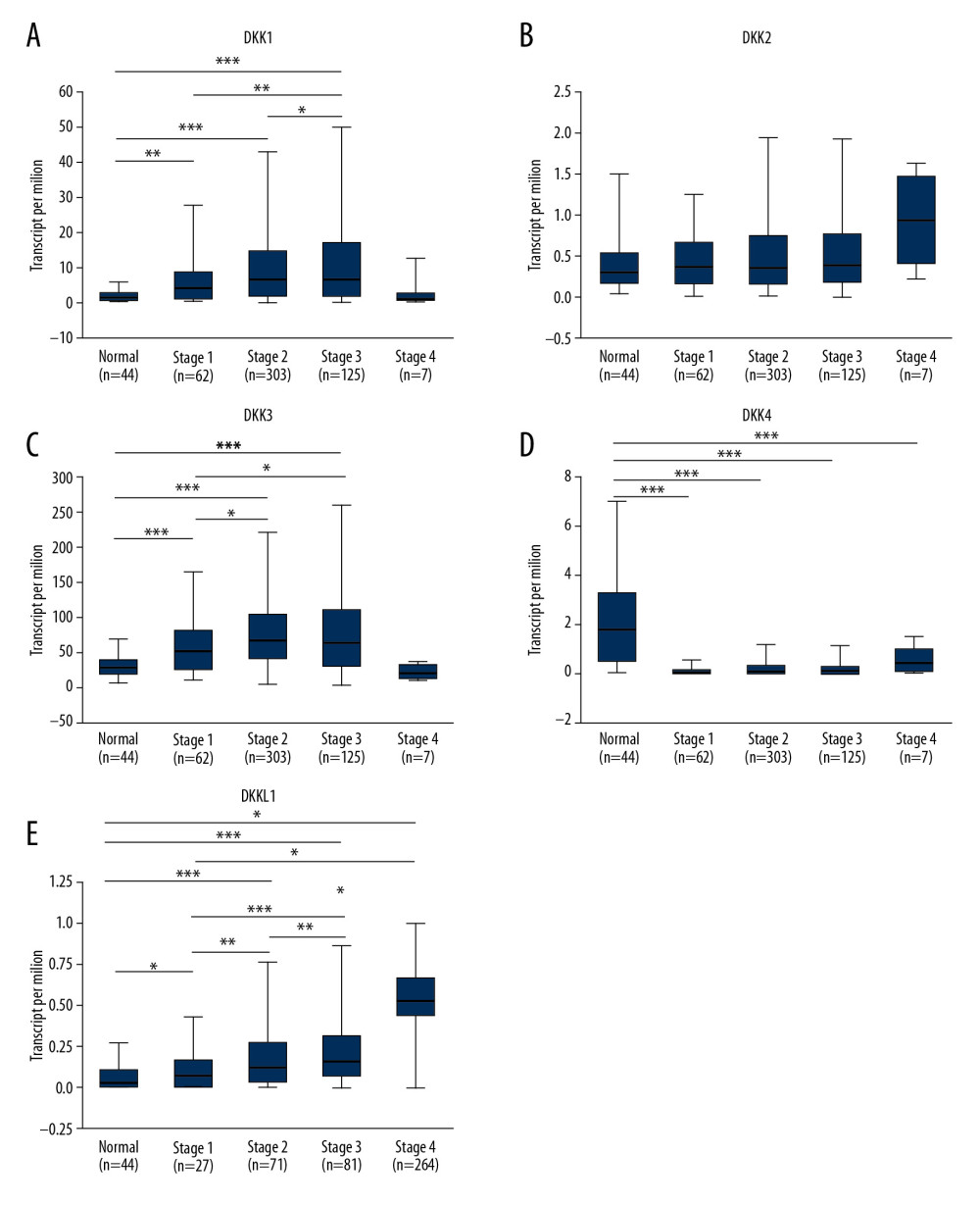 Figure 4. (A–E) Association of the mRNA expression of distinct DKKs and tumor grades of HNSCC patients. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 4. (A–E) Association of the mRNA expression of distinct DKKs and tumor grades of HNSCC patients. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. ![(A–E) Association of the mRNA expression of distinct DKKs with the HPV status of HNSCC patients. HPV status and viral subtypes were assessed by DNA sequencing and PathSeq algorithm [20]. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma; HPV – human papillomavirus.](https://jours.isi-science.com/imageXml.php?i=medscimonit-26-e927368-g005.jpg&idArt=927368&w=1000) Figure 5. (A–E) Association of the mRNA expression of distinct DKKs with the HPV status of HNSCC patients. HPV status and viral subtypes were assessed by DNA sequencing and PathSeq algorithm [20]. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma; HPV – human papillomavirus.
Figure 5. (A–E) Association of the mRNA expression of distinct DKKs with the HPV status of HNSCC patients. HPV status and viral subtypes were assessed by DNA sequencing and PathSeq algorithm [20]. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma; HPV – human papillomavirus. 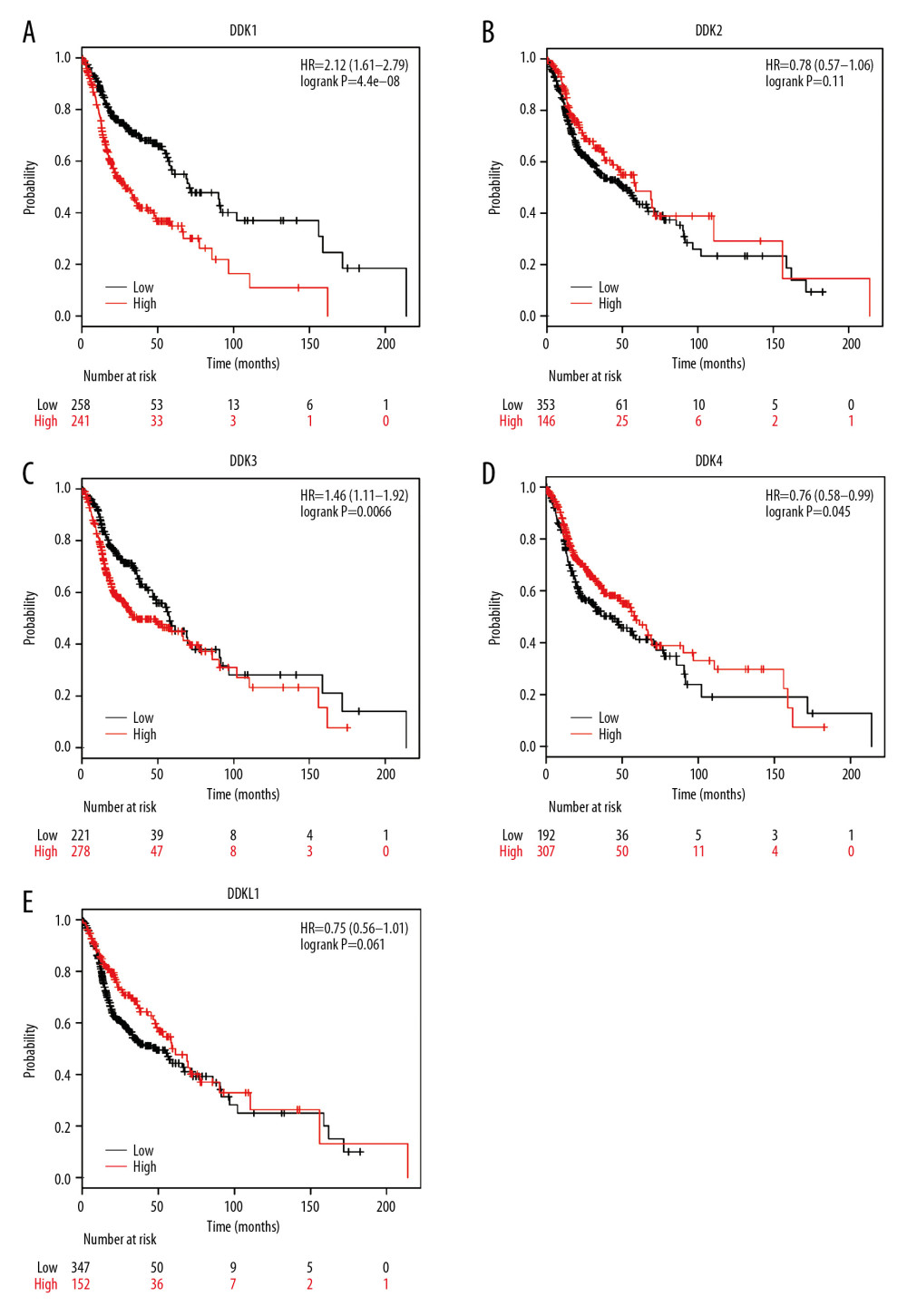 Figure 6. (A–E) Prognostic value of the mRNA expression of distinct DKKs in HNSCC patients (Kaplan-Meier Plotter). DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 6. (A–E) Prognostic value of the mRNA expression of distinct DKKs in HNSCC patients (Kaplan-Meier Plotter). DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. 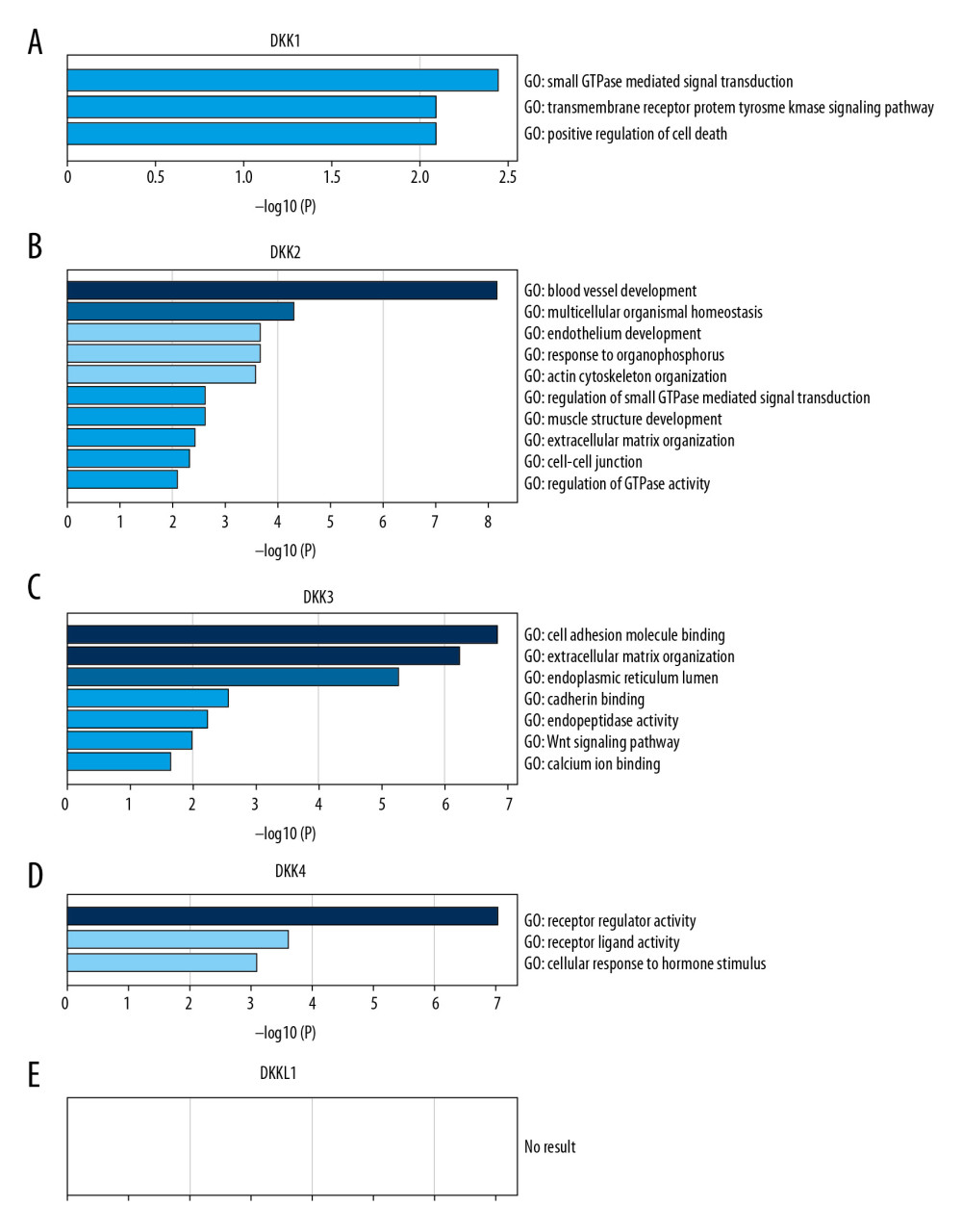 Figure 7. (A–E) Possible functions and pathways of distinct DKKs and the top 20 similar genes in HNSCC. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 7. (A–E) Possible functions and pathways of distinct DKKs and the top 20 similar genes in HNSCC. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. 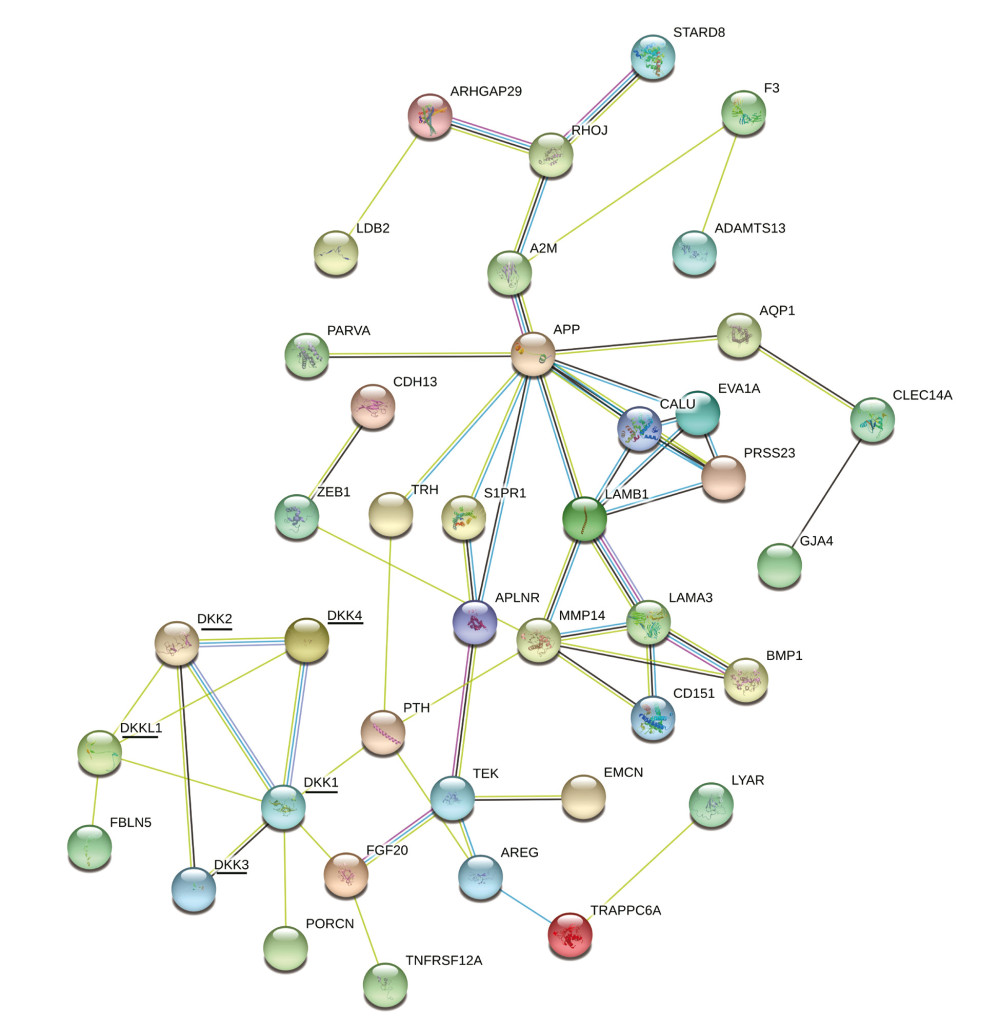 Figure 8. PPI network of all DKKs and their similar genes. PPI – protein–protein interaction; DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 8. PPI network of all DKKs and their similar genes. PPI – protein–protein interaction; DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. References
1. Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019: Cancer J Clin, 2019; 69; 7-34
2. Marur S, Forastiere AA, Head and neck cancer: Changing epidemiology, diagnosis, and treatment: Mayo Clin Proc, 2008; 83; 489-501
3. Gooi Z, Chan JY, Fakhry C, The epidemiology of the human papillomavirus related to oropharyngeal head and neck cancer: Laryngoscope, 2016; 126; 894-900
4. Chan JYK, Zhen G, Agrawal N, The role of tumor DNA as a diagnostic tool for head and neck squamous cell carcinoma: Semin Cancer Biol, 2019; 55; 1-7
5. Niehrs C, Function and biological roles of the Dickkopf family of Wnt modulators: Oncogene, 2006; 25; 7469-81
6. Igbinigie E, Guo F, Jiang SW, Dkk1 involvement and its potential as a biomarker in pancreatic ductal adenocarcinoma: Clin Chim Acta, 2019; 488; 226-34
7. Katase N, Gunduz M, Beder LB, Frequent allelic loss of Dkk-1 locus (10q11.2) is related with low distant metastasis and better prognosis in head and neck squamous cell carcinomas: Cancer Invest, 2010; 28; 103-10
8. Hamzehzadeh L, Caraglia M, Atkin SL, Sahebkar A, Dickkopf homolog 3 (DKK3): A candidate for detection and treatment of cancers?: J Cell Physiol, 2018; 233; 4595-605
9. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles: Neoplasia, 2007; 9; 166-80
10. Chandrashekar DS, Bashel B, Balasubramanya SAH, UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses: Neoplasia, 2017; 19; 649-58
11. Nagy Á, Lánczky A, Menyhárt O, Győrffy B, Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets: Sci Rep, 2018; 8; 9227
12. Tang Z, Li C, Kang B, GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses: Nucleic Acids Res, 2017; 45; W98-102
13. Zhou Y, Zhou B, Pache L, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets: Nat Commun, 2019; 10; 1523
14. Szklarczyk D, Gable AL, Lyon D, STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets: Nucleic Acids Res, 2019; 47; D607-13
15. Peng CH, Liao CT, Peng SC, A novel molecular signature identified by systems genetics approach predicts prognosis in oral squamous cell carcinoma: PLoS One, 2011; 6; e23452
16. Toruner GA, Ulger C, Alkan M, Association between gene expression profile and tumor invasion in oral squamous cell carcinoma: Cancer Genet Cytogenet, 2004; 154; 27-35
17. Ginos MA, Page GP, Michalowicz BS, Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck: Cancer Res, 2004; 64; 55-63
18. Pyeon D, Newton MA, Lambert PF, Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers: Cancer Res, 2007; 67; 4605-19
19. Sengupta S, den Boon JA, Chen IH, Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma: Cancer Res, 2006; 66; 7999-8006
20. Campbell JD, Yau C, Bowlby R, Genomic, pathway network, and immunologic features distinguishing squamous carcinomas: Cell Rep, 2018; 23; 194-212.e6
21. Huang Y, Liu L, Liu A, Dickkopf-1: Current knowledge and related diseases: Life Sci, 2018; 209; 249-54
22. Lee AY, He B, You L, Dickkopf-1 antagonizes Wnt signaling independent of beta-catenin in human mesothelioma: Biochem Biophys Res Commun, 2004; 323; 1246-50
23. Cho SW, Lee EJ, Kim H, Dickkopf-1 inhibits thyroid cancer cell survival and migration through regulation of beta-catenin/E-cadherin signaling: Mol Cell Endocrinol, 2013; 366; 90-98
24. Chen L, Li M, Li Q, DKK1 promotes hepatocellular carcinoma cell migration and invasion through β-catenin/MMP7 signaling pathway: Mol Cancer, 2013; 12; 157
25. Gao H, Li L, Xiao M, Elevated DKK1 expression is an independent unfavorable prognostic indicator of survival in head and neck squamous cell carcinoma: Cancer Manag Res, 2018; 10; 5083-89
26. Kawakita A, Yanamoto S, Yamada S, MicroRNA-21 promotes oral cancer invasion via the Wnt/beta-catenin pathway by targeting DKK2: Pathol Oncol Res, 2014; 20; 253-61
27. Yang J, Jiang Y, He R, DKK2 impairs tumor immunity infiltration and correlates with poor prognosis in pancreatic ductal adenocarcinoma: J Immunol Res, 2019; 2019 8656282
28. Katase N, Nishimatsu SI, Yamauchi A, DKK3 overexpression increases the malignant properties of head and neck squamous cell carcinoma cells: Oncol Res, 2018; 26; 45-58
29. Katase N, Lefeuvre M, Tsujigiwa H, Knockdown of Dkk-3 decreases cancer cell migration and invasion independently of the Wnt pathways in oral squamous cell carcinoma-derived cells: Oncol Rep, 2013; 29; 1349-55
30. Yin DT, Wu W, Li M, DKK3 is a potential tumor suppressor gene in papillary thyroid carcinoma: Endocr Relat Cancer, 2013; 20; 507-14
31. Cai X, Yao Z, Li L, Huang J, Role of DKK4 in tumorigenesis and tumor progression: Int J Biol Sci, 2018; 14; 616-21
32. Tarnowski M, Czerewaty M, Deskur A, Expression of cancer testis antigens in colorectal cancer: New prognostic and therapeutic implications: Dis Markers, 2016; 2016 1987505
Figures
 Figure 1. The mRNA expression of DKKs in 20 different types of cancers (Oncomine database). The figure showed the numbers of datasets with statistically significant mRNA overexpression (red) or downregulated expression (blue) of DKKs. The difference in transcriptional expression was analyzed by t test. Cut-off of P value and fold change were as follows: P=0.01; fold change, 1.5; gene rank, 10%; data type, mRNA. DKK – Dickkopf Wnt signaling pathway inhibitor.
Figure 1. The mRNA expression of DKKs in 20 different types of cancers (Oncomine database). The figure showed the numbers of datasets with statistically significant mRNA overexpression (red) or downregulated expression (blue) of DKKs. The difference in transcriptional expression was analyzed by t test. Cut-off of P value and fold change were as follows: P=0.01; fold change, 1.5; gene rank, 10%; data type, mRNA. DKK – Dickkopf Wnt signaling pathway inhibitor. Figure 2. (A–E) The mRNA expressions of distinct DKKs in HNSCC tissues and normal controls (UALCAN). *** P<0.001. DKK, Dickkopf Wnt signaling pathway inhibitor; HNSCC, head and neck squamous cell carcinoma.
Figure 2. (A–E) The mRNA expressions of distinct DKKs in HNSCC tissues and normal controls (UALCAN). *** P<0.001. DKK, Dickkopf Wnt signaling pathway inhibitor; HNSCC, head and neck squamous cell carcinoma. Figure 3. (A–E) Relationship between the mRNA expression of distinct DKKs and tumor stage of HNSCC patients. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 3. (A–E) Relationship between the mRNA expression of distinct DKKs and tumor stage of HNSCC patients. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. Figure 4. (A–E) Association of the mRNA expression of distinct DKKs and tumor grades of HNSCC patients. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 4. (A–E) Association of the mRNA expression of distinct DKKs and tumor grades of HNSCC patients. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.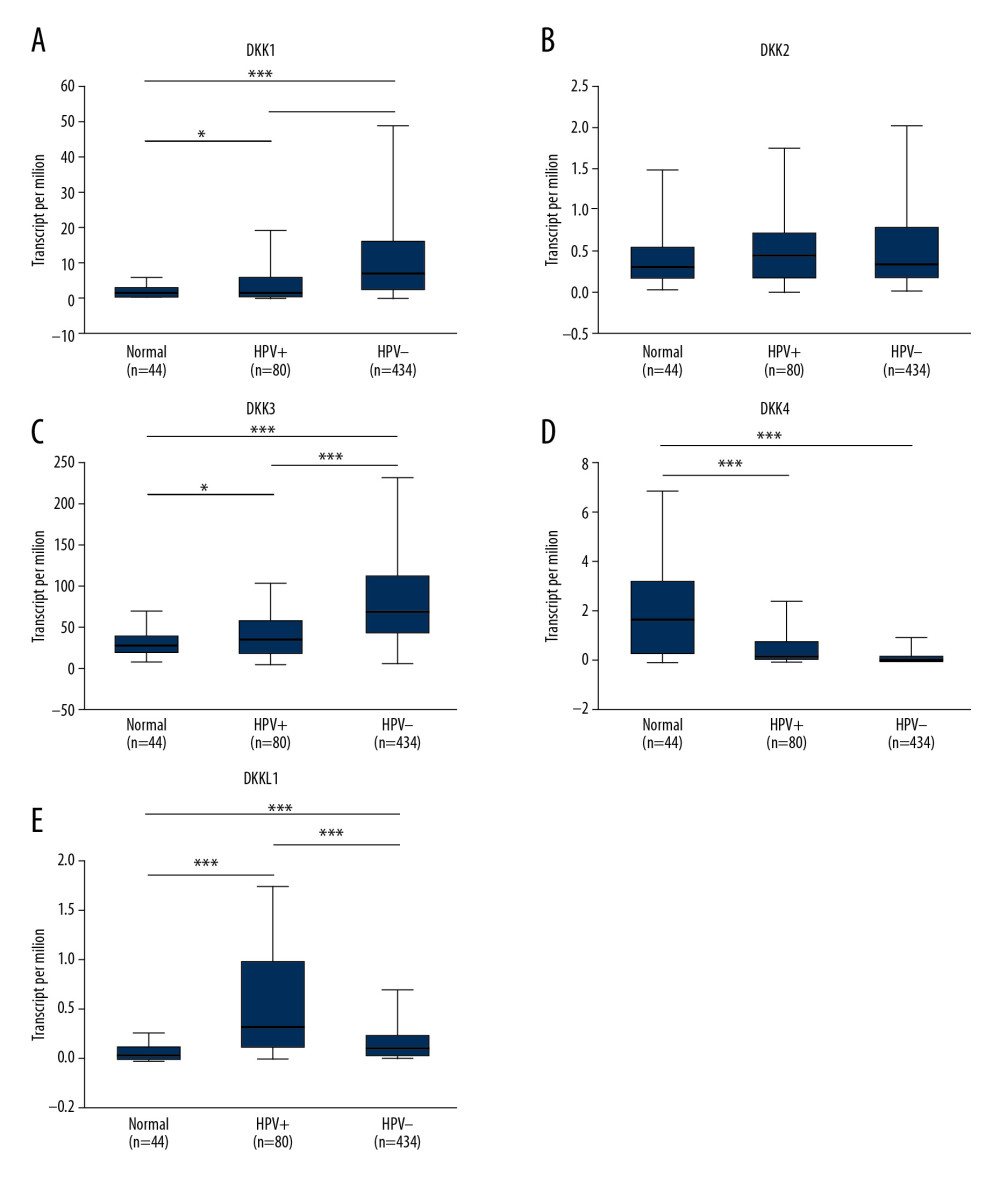 Figure 5. (A–E) Association of the mRNA expression of distinct DKKs with the HPV status of HNSCC patients. HPV status and viral subtypes were assessed by DNA sequencing and PathSeq algorithm [20]. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma; HPV – human papillomavirus.
Figure 5. (A–E) Association of the mRNA expression of distinct DKKs with the HPV status of HNSCC patients. HPV status and viral subtypes were assessed by DNA sequencing and PathSeq algorithm [20]. * P<0.05, ** P<0.01, *** P<0.001. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma; HPV – human papillomavirus. Figure 6. (A–E) Prognostic value of the mRNA expression of distinct DKKs in HNSCC patients (Kaplan-Meier Plotter). DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 6. (A–E) Prognostic value of the mRNA expression of distinct DKKs in HNSCC patients (Kaplan-Meier Plotter). DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. Figure 7. (A–E) Possible functions and pathways of distinct DKKs and the top 20 similar genes in HNSCC. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 7. (A–E) Possible functions and pathways of distinct DKKs and the top 20 similar genes in HNSCC. DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. Figure 8. PPI network of all DKKs and their similar genes. PPI – protein–protein interaction; DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma.
Figure 8. PPI network of all DKKs and their similar genes. PPI – protein–protein interaction; DKK – Dickkopf Wnt signaling pathway inhibitor; HNSCC – head and neck squamous cell carcinoma. In Press
05 Mar 2024 : Clinical Research
Muscular Function Recovery from General Anesthesia in 132 Patients Undergoing Surgery with Acceleromyograph...Med Sci Monit In Press; DOI: 10.12659/MSM.942780
05 Mar 2024 : Clinical Research
Effects of Thermal Insulation on Recovery and Comfort of Patients Undergoing Holmium Laser LithotripsyMed Sci Monit In Press; DOI: 10.12659/MSM.942836
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952