05 February 2021: Clinical Research
Effects of Antibiotic Treatment and Probiotics on the Gut Microbiome of 40 Infants Delivered Before Term by Cesarean Section Analysed by Using 16S rRNA Quantitative Polymerase Chain Reaction Sequencing
Chen Gong1DEF, Liqi Yang1G, Kangkang Liu1C, Shichun Shen2F, Qixing Zhang1B, Han Li1B, Yan Cheng1A*DOI: 10.12659/MSM.928467
Med Sci Monit 2021; 27:e928467
Abstract
BACKGROUND: This study aimed to investigate the effects on the gut microbiome of 40 infants delivered before term by cesarean section between antibiotic treatment and probiotics as assessed by 16S rRNA quantitative polymerase chain reaction (qPCR) sequencing.
MATERIAL AND METHODS: We divided 40 premature infants delivered by cesarean section into 4 groups according to exposure to antibiotics or probiotics: N group (No-probiotics and No-antibiotics), A group (antibiotics), P group (probiotics), and the AP group (antibiotics+probiotics). Fecal samples were collected on days 1, 3, and 10, and the microflora data were generated using 16S rRNA qPCR sequencing technology. The BugBase tool was used for phenotype prediction, the Tax4Fun tool was used for function prediction, and iPath software was used to predict the metabolic pathways of intestinal bacteria.
RESULTS: Antibiotics increased the abundance of pathogenic bacteria and reduced the replication and repair function (P=0.049), nucleotide metabolism function (P=0.047), and the purine metabolism pathways (P<0.05) of the gut microbiota. Probiotics increased the abundance of beneficial bacteria and the cellular community prokaryote function (P=0.042) and contributed to the Bifidobacteria biofilm formation. Probiotics alleviated the damage of antibiotics to the composition and function of the gut microbiota.
CONCLUSIONS: The findings from this study showed that antibiotic treatment of preterm infants born by cesarean section changed the gut microbiome, but that the use of probiotics could restore the normal microbiome, which supports that restoration of the normal gut microbiota may be achieved with probiotics.
Keywords: Cesarean Section, Infant, Premature, Antibiotic Prophylaxis, Probiotics, Anti-Bacterial Agents, Bacteria, Feces, Infant, Infant, Newborn, Polymerase Chain Reaction
Background
The human gut microbiota, which is composed of complex microbial communities, is a complex ecosystem shaped by evolution, where the combination of host and bacteria promote a delicate balance evolved to modulate immune responses and promote health. Understanding the process of colonization by gastrointestinal microorganisms involves defining host physiology and immunity, and the neonatal period is the essential and susceptible stage in this process [1].
Premature infants are born between 28 and 37 weeks of gestational age. Compared with full-term infants, the intestinal microflora of premature infants is established significantly later, the abundance and diversity of microflora are lower, and there are more types and contents of pathogenic bacteria [2,3]. However, the gut microbiota of premature infants is characterized by low microbial diversity and high inter-individual variation [4]. Many studies have indicated that this discrepancy might be caused by frequent exposure of preterm infants to various medical treatments [5,6]; delivery mode and use of antibiotics are the most common of these [2,7]. Cesarean section decreases the abundance of
Many studies have demonstrated that sepsis, NEC, and feeding intolerance are closely related to gut microbiota imbalance in premature infants [14–16]. Furthermore, brain injury in premature infants and nervous system development are related to gut microbiota disorders [17]. Hence, it is important to intervene in the gut microbiota of premature infants as early as possible. Use of probiotics is currently the most commonly used intervention method. Probiotics increase beneficial bacteria and decrease pathogenic bacteria in preterm infants [18], and can reduce the risk of NEC and feeding intolerance in preterm infants, as well as reducing the expression level of inflammatory factors to minimize brain damage [19,20]. Research has shown that long-term probiotic supplementation can help restore normal microbiota in 3-month-old antibiotic-treated infants [21].
However, previous studies did not deeply explore the effects of probiotics supplementation in early life on the gut flora of infants delivered before term by cesarean section and treated with antibiotics. The 16S rRNA quantitative polymerase chain reaction (qPCR) sequencing method is mainly used to identify strains through sequencing and analysis of the specific 16S rRNA high-variation region. It is an important technology for the study of the microbial community and identifies broad levels of community composition. Therefore, the present prospective study aimed to investigate the effects on the microbiome of 40 infants delivered before term by cesarean section between antibiotic treatment and probiotics by use of 16S rRNA quantitative PCR sequencing.
Material and Methods
PARTICIPANT SELECTION:
In this study, 40 premature infants born by cesarean section at 28 to 37 weeks of gestational age were recruited with parental informed consent. Among the 40 premature infants, 20 premature infants had high-risk factors for infection (eg, premature rupture of membranes over 18 h, inhaled contaminated amniotic fluid or meconium during labor, and clinical manifestations suspected of early infection) and needed antibiotic treatment based on the clinical guidelines for the use of antibiotics. There were 20 premature infants without infectious factors and without antibiotic treatment. This study was conducted in strict accordance with the Helsinki Declaration and was approved by the Ethics Committee of the Second Affiliated Hospital of Anhui Medical University (approval no. Pj2016-05-01).
Exclusion criteria were: severe asphyxia at birth, neonatal respiratory distress syndrome (NRDS), genetic metabolic diseases, congenital pyloric obstruction, congenital megacolon, and other digestive tract malformations.
EXPERIMENTAL DESIGN:
Twenty premature infants treated with antibiotics were randomized to 2 groups, one group with probiotics and the other group without probiotics (10 cases in each group): the AP group (antibiotics + probiotics) and the A group (antibiotics). Another 20 preterm infants without antibiotic treatment were also randomized to 2 groups, one with probiotics and the other without probiotics (10 cases in each group): the P group (probiotics) and the N group (No-probiotics and No-antibiotics).
Antibiotic usage method: In this study, Cefotaxime (North China Pharmaceutical Hebei Huamin Pharmaceutical Company limited, H10960279) was selected. Premature infants of the AP group and A group were given antibiotic treatment within 24 h of birth. The antibiotic dosage was 50 milligrams per kilogram of body weight, twice daily, and the duration of therapy was 7 days.
Probiotics usage method: The probiotic was Bifid triple viable powder (Shanghai Shangyao Xinyi Pharmaceutical Factory Company limited, S10970104). One gram of the probiotic powder contains a mixture of cells (quantified as colony-forming units, CFU) of
To exclude the influence of feeding mode, the premature infants in this study were all fed with formula milk.
STOOL SPECIMEN COLLECTION:
Fecal samples from the 4 groups of premature infants were collected on days 1, 3, and 10. Day 10 fecal specimens were samples after 7 days of exposure to probiotics or antibiotics. The fecal samples were immediately stored in a sterilized frozen test tube, which was transferred to the refrigerator at −80°C in liquid nitrogen for preservation.
GENOME DNA EXTRACTION AND PCR AMPLIFICATION: The genomic DNA of stool samples was extracted by the cetyl trimethyl ammonium bromide (CTAB) method as described by Tang et al [22]. We used 1% agarose gel to detect the concentration and purity of DNA. The genomic DNA diluted to 1 ng/uL with sterile water was then used as a template. The corresponding primer (16S V3–V4: 341F-806R, 5′-CCTAYGGGRBGCASCAG and 5′-GGACTACNNGGGTATCTAAT) with the barcode were selected to amplify the 16S rRNA genes of regions (16S V3–V4). All PCR reactions were carried out in 30 μL reactions with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs); 0.2 μM of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, followed by a final cycle of 72°C for 5 min. We mixed the same volume of 1×loading buffer (contained SYB green) with PCR products and performed electrophoresis on 2% agarose gel for detection. PCR products were mixed in equidensity ratios. Then, the mixture of PCR products was purified using a GeneJETTM Gel Extraction Kit (Thermo Scientific) [23]. The target bands were cut and recovered after PCR products were mixed and purified. Then, the library was built and sequenced on the IonS5TMXL sequencing platform (ThermoFisher Scientific, Waltham, MA, USA).
OPERATIONAL TAXONOMIC UNITS (OTUS) CLUSTER AND SPECIES ANNOTATION:
Based on the effective data obtained by quality control, filtering, and chimera removal of the original sequencing results, Uparse software (Uparse v7.0.1001) was used to screen representative sequences for each OTU. Then, the Silva Database was used based on the Mothur algorithm to annotate taxonomic information for the representative sequences. Finally, we standardized the abundance information of the OTU and performed data analysis.
EXPERIMENTAL DATA ANALYSIS:
QIIME (version 1.7.0) was used to calculate alpha diversity index, including the abundance-based coverage estimator (ACE) and Chao1, and the results were displayed by R software (Version 2.15.3). Spearman correlation analysis was performed using vegdist function and mantel function in the Vegan package in R. In addition, the BugBase tool was used for phenotype prediction of intestinal flora, the Tax4Fun tool was used to predict the function of gut microbiota, and iPath software was used to predict the metabolic pathways of intestinal bacteria.
SPSS 19.0 (Version SPSS Inc., Chicago, IL, USA) was used to analyze the basic information of the samples. The statistical significance of differences was evaluated by one-way ANOVA (SNK method) and chi-square test. The results were expressed as the mean±SD.
Results
PARTICIPANT CHARACTERISTICS:
A total of 40 premature infants were enrolled in this study, all of whom were delivered by cesarean section. A total of 120 stool samples were collected. SPSS 19.0 software was used for statistical analysis of the basic information of the premature infants in the 4 groups, and the results indicated that there were no significant differences among groups in birth weight, gestational age, and sex (P>0.05) (Table 1).
CHARACTERISTIC OF GUT MICROBIOTA IN PREMATURE INFANTS DELIVERED BY CESAREAN SECTION:
After 16S rRNA sequencing, a total of 10 878 species of OTU were found in all the fecal specimens of the premature infants, and 57 phyla and 1058 genera were identified. At the phylum level, the gut microbiota of all of the premature infants delivered by cesarean section was mainly composed of Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, accounting for more than 90% of the microorganisms in the fecal samples. At the phylum level, the gut microbiota of the 4 groups of premature infants showed a trend of decreasing Proteobacteria and increasing Firmicutes over time (days 1, 3, and 10), and Firmicutes were dominant at day 10 (Figure 1).
DIFFERENCES IN COMPOSITION OF THE GUT MICROBIOTA ON DAY10:
Since the premature infants had been exposed to probiotics or antibiotics for 7 days at day10, we compared the effects of antibiotics or probiotics at day 10 on the composition of the gut microbiota. At the genus level, in the A-day10 group, the abundance of unidentified Enterobacteriaceae and Parabacteroides was higher, whereas the abundance of Bifidobacterium was lower than in the N-day10 group. Compared with the N-day10 group, the abundance of Bifidobacterium, Lactobacillus, and Enterococcus were higher in the P-day10 group (Figure 2A).
To discuss the effect of probiotics on the composition in the gut microbiota of premature infants delivered by cesarean section treated with antibiotics, LEfSe (linear discriminant analysis) was used to analyze the biomarkers (the major bacteria causing the difference between the 2 groups) of groups AP-day10 and A-day10. The results indicated that 24 biomarkers (order: 2, family: 8, genus: 8, species: 6) were found, and the biomarkers of the AP-day10 group primarily contained Lactobacillus (genus level), Bifidobacterium bifidum, and Bifidobacterium animals (species level), among others. The A-day10 group mainly included unidentified Enterobacteriaceae, unidentified Clostridiales (genus level), Escherichia coli, Clostridium paraputrificum, and Clostridium difficile (species level) (Figure 2B).
ANALYSIS OF THE DIVERSITY OF THE GUT MICROBIOTA:
The ACE index and Chao1 index were used to evaluate the alpha diversity of the gut microbiota on day 10 in the 4 groups of preterm infants delivered by cesarean section. The results showed that there were no statistically significant differences in the ACE index and Chao1 index values between the N-day10 group and the A-day10 group (P>0.05). Compared with the N-day10 group, the ACE index and Chao1 index of the P-day10 group were significantly lower (P<0.01). Similarly, the ACE and Chao1 indexes in the AP-day10 group were lower than in the A-day10 group (P<0.01) (Figure 3A, 3B).
CORRELATION ANALYSIS OF ENVIRONMENTAL FACTORS:
To study the influence of environmental factors on the gut flora of premature infants delivered by cesarean section, the Spearman method was used to analyze the relationship between the gut microbiota and 5 environmental factors: birth weight, gestational age, blood albumin (Alb), blood lactate, and blood pH at birth.
Experimental results show that, at the genus level, the gut microbiota of premature infants was mainly significantly correlated with Alb. Stenotrophomonas (P=0.03, r=−0.47) and Sphingomonas (P=0.01, r=−0.53) were negatively correlated with Alb. Rothia (P=0.04, r=0.45) and Clostridioides (P=0.01, r=0.52) were positively correlated with Alb (Figure 4).
PHENOTYPIC PREDICTION OF THE GUT MICROBIOTA:
BugBase was used to predict the phenotype of gut microbiota. We found that the time trend of intestinal microecology of the 4 groups of premature infants from days 1 to 3 to 10 showed a gradual decrease in the abundance of aerobic bacteria, and there were significant differences between day 1 and day 10 in the 4 groups (P<0.05) (Figure 5A). There was a gradual increase in the abundance of anaerobic bacteria, but there were no significant differences between day 1 and day 10 in the 4 groups (P>0.05) (Figure 5B).
Figure 5C shows the relative abundance diagram of biofilm-forming bacteria in each group. It was found that the biofilm formation in groups N-day10 and A-day10 was mainly Proteobacteria, and the biofilm formation in groups P-day10 and AP-day10 using probiotics was Proteobacteria and Actinobacteria (mainly Bifidobacteria) (Figure 5C).
FUNCTION PREDICTION ANALYSIS OF INTESTINAL FLORA:
Tax4Fun software was used to predict the function of the gut microbiota, and the functions with significant differences among the groups were compared (P<0.05). The results of function prediction showed that the replication and repair (P=0.049) and nucleotide metabolism (P=0.047) of the N-day10 group were significantly higher than in the A-day10 group (Figure 6A). The cellular community Prokaryotes (P=0.042) of the P-day10 group were significantly higher than in the N-day10 group (Figure 6B). In addition, compared with the A-day10 group, replication and repair (P=0.01) and nucleotide metabolism (P=0.001) of the AP-day10 group were significantly higher, whereas the cardiovascular disease (P=0.009) was lower (Figure 6C).
METABOLIC PATHWAY PREDICTION ANALYSIS:
Given the differences in nucleotide metabolism function between groups N-day10 and A-day10, the metabolism function of the 2 groups was further investigated. The approach was based on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologous groups extracted from fecal samples of groups N-day10 and A-day10 to predict and map metabolic pathways. The results showed that the purine metabolism (map00230), one-carbon pool by folate (map00670), photosynthesis (map00195), and carbon fixation pathways in Prokaryotes (map00720) of the A-day10 group were significantly lower than in the N-day10 group (Figure 7).
Discussion
In this study, we investigated the composition and metabonomics of gut microbiota in premature infants delivered by cesarean section using 16S rRNA Quantitative Polymerase Chain Reaction Sequencing techniques. The results showed that early application of antibiotics significantly changed the gut microbiota, antibiotics reduced the abundance of
The gut microbiota of premature infants born by cesarean section was mainly composed of 4 significant phyla, which is consistent with the literature [24]. In addition, the variational tendency from
On day 10, the composition of the gut microbiota showed differences in abundance at the genus level. Compared with the N-day10 group, the A-day10 group receiving antibiotics had a higher abundance of unidentified
LEfSe was used to analyze the biomarkers of AP-day10 and A-day10 groups, so as to assess the effect of probiotics on the gut microbiota of premature infants delivered by cesarean section treated with antibiotics. The results showed that the AP-day10 group contained more beneficial bacteria (
In the analysis of gut microbiota diversity, a previous study showed no significant changes in the gut microbial diversity at week 4 for term infants given early, short-term antibiotic treatment [12]. A study has also shown that antibiotic exposure in the first few days after birth did not significantly change the diversity of the gut microbiota in premature infants [32]. Therefore, there was no significant difference in the gut microbiota diversity of antibiotic-treated premature infants delivered by cesarean section, which may be due to the short duration of antibiotics use. Consistent with previous studies, the 2 groups of preterm infants treated with probiotics in this experiment had significantly lower alpha diversity of gut flora, which may account for the decrease in the number of harmful bacteria [18].
Correlation analysis of environmental factors showed that, at the genus level, Alb was the major factor affecting the composition of the gut microbiota, and the
The phenotype prediction analysis showed that the intestinal microecology of premature infants delivered by cesarean section showed a trend of transformation from aerobic bacteria to anaerobic bacteria over time. This may be because the gut is rich in oxygen at birth, when it is suitable for aerobic bacteria to grow, and anaerobic bacteria begin to grow in large quantities after oxygen is used up. In addition, biofilm formation of
The results of function prediction found that both antibiotics and probiotics had significant effects on the gut microbiota function of premature infants delivered by cesarean section. A study on the gut microbiota of mice treated with antibiotics revealed that the use of antibiotics decreases the translation, replication, recombination, and repair functions [39]. Similar to the gut microbiota experiment in mice, the replication and repair function in preterm infants delivered by cesarean section receiving antibiotics (A-day10 group) was also significantly reduced. In addition, the nucleotide metabolism function of the A-day10 group was significantly lower than that of the N-day10 group in this experiment, indicating the adverse effect of the antibiotics on the gut microbiota function of preterm infants in cesarean section. Regarding the effects of probiotics, the cellular community of
The results of metabolic pathway prediction in N-day10 and A-day10 groups showed that the purine metabolic pathway and one-carbon pool by folate pathway were significantly lower in cesarean section preterm infants treated with antibiotics compared with that in the N-day10 group. A previous study showed that the gut microbiota is involved in purine metabolism, and abnormal purine metabolism may increase uric acid [43]. In this study, the purine metabolism pathway in group A-day10 was lower than that in group N-day10, which may be due to the disorder of gut microbiota caused by using antibiotics, thus affecting the purine metabolism pathway. The differences in purine metabolic pathways may be the reason that the nucleotide metabolism function of the A-day10 group was lower than in the N-day10 group. In addition, the one-carbon pool by folate pathway in group A-day10 was also significantly lower than in the N-day10 group. Folic acid is one of the essential B vitamins involved in DNA synthesis and repair [9]. In this experiment, the replication and repair function of the A-day10 group was lower than that of the N-day10 group. We speculate that this function might be affected by the abnormal metabolic pathway. Therefore, the use of antibiotics has an adverse effect on some metabolic pathways of intestinal flora in premature infants delivered by cesarean section.
To the best of our knowledge, the present study is the first to indicate that probiotics supplementation can restore the composition and function of gut flora in antibiotic-treated premature infants delivered by cesarean section. However, there are some limitations to our research that need to be considered. For example, although the high-throughput sequencing of 16S rRNA is currently the most widely used technique in analysis of the microbial community, it is limited in resolution compared with metagenomics [44]. In addition, to ensure the accuracy of the experiment, all the 40 premature infants by cesarean section selected in this study were fed with formula milk, which may ignore the effect of breastfeeding on gut flora. Accordingly, future studies should assess the effects of probiotics or antibiotics on intestinal flora of breastfed premature infants delivered by cesarean section. Metagenomics and proteomics may also be combined to further explore this topic in future studies.
Conclusions
In conclusion, the findings from this study showed that antibiotic treatment of preterm infants born by cesarean section changed the composition and function of gut microbiome, but that the use of probiotics could restore the normal microbiome, which supports that restoration of the normal gut microbiota may be achieved with probiotics. Furthermore, the relationship between
Figures
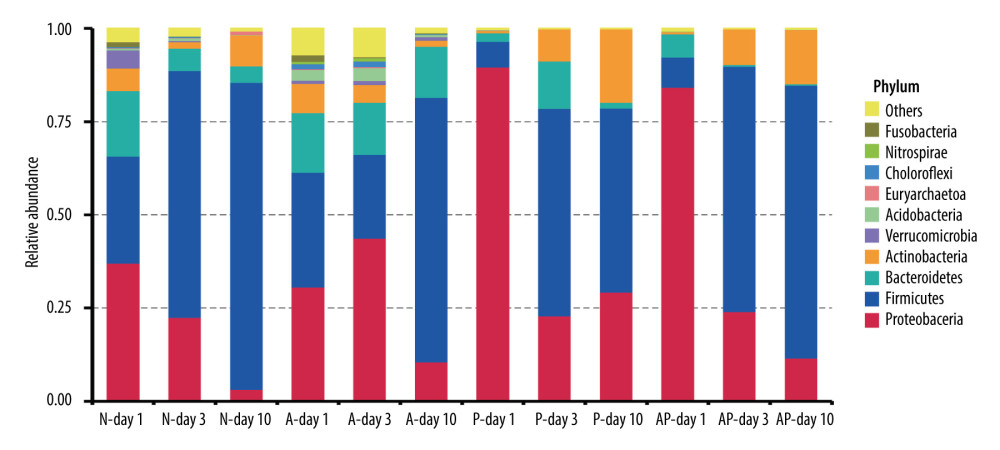 Figure 1. The composition of gut microbiome of preterm infants delivered by cesarean section in the 4 groups at 1, 3, and 10 days: Ten most abundant bacteria at the phylum level. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day1 – One day after birth; day3 – Three days after birth; day10 – Ten days after birth.
Figure 1. The composition of gut microbiome of preterm infants delivered by cesarean section in the 4 groups at 1, 3, and 10 days: Ten most abundant bacteria at the phylum level. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day1 – One day after birth; day3 – Three days after birth; day10 – Ten days after birth. 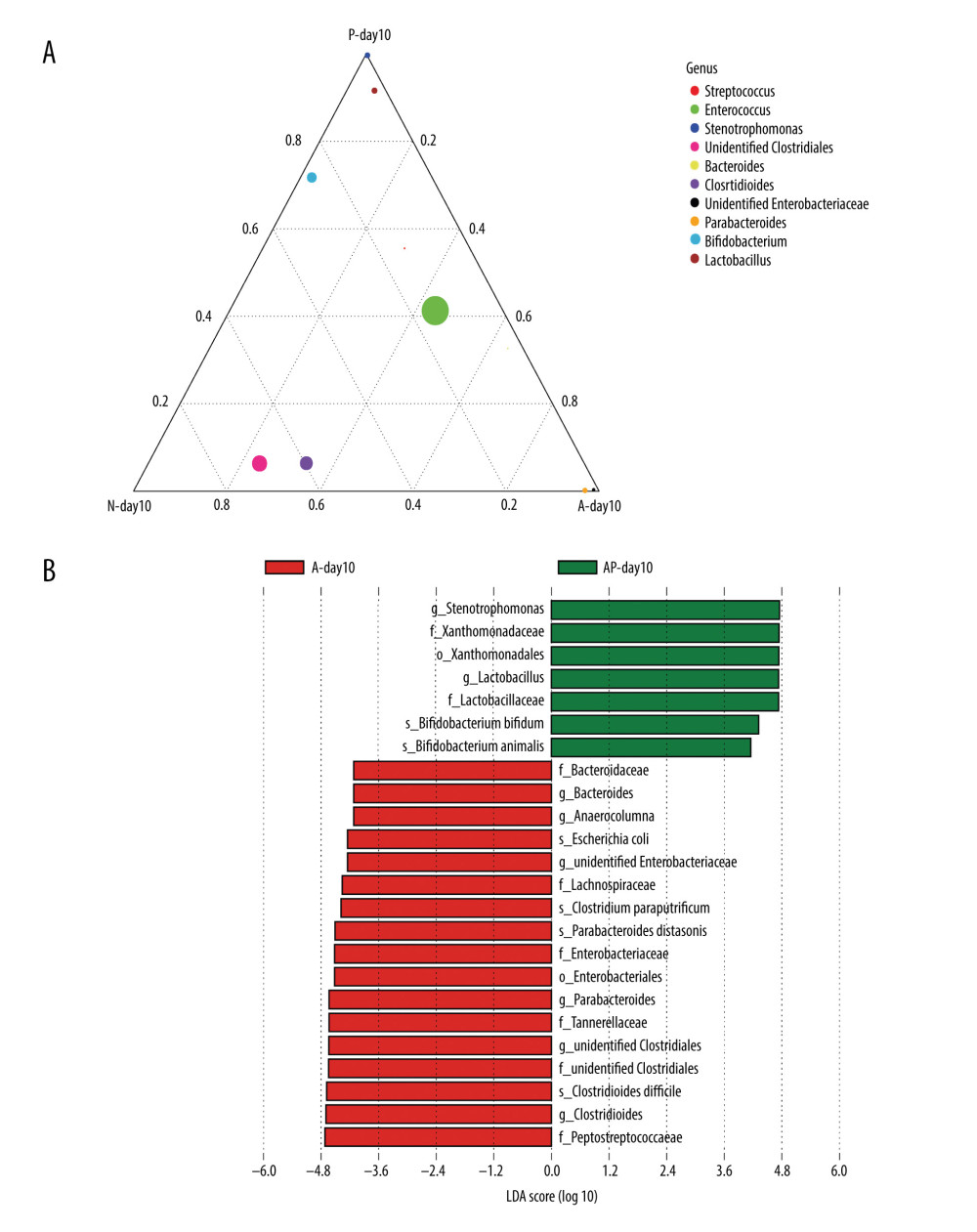 Figure 2. Differences of gut microbiome composition of preterm infants delivered by cesarean section in the 4 groups at 10 days. (A) Differences in N-day10, A-day10 and P-day10 group at the genus level. (B) Significance difference Biomarker between the A-day10 and AP-day10 groups. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth.
Figure 2. Differences of gut microbiome composition of preterm infants delivered by cesarean section in the 4 groups at 10 days. (A) Differences in N-day10, A-day10 and P-day10 group at the genus level. (B) Significance difference Biomarker between the A-day10 and AP-day10 groups. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. 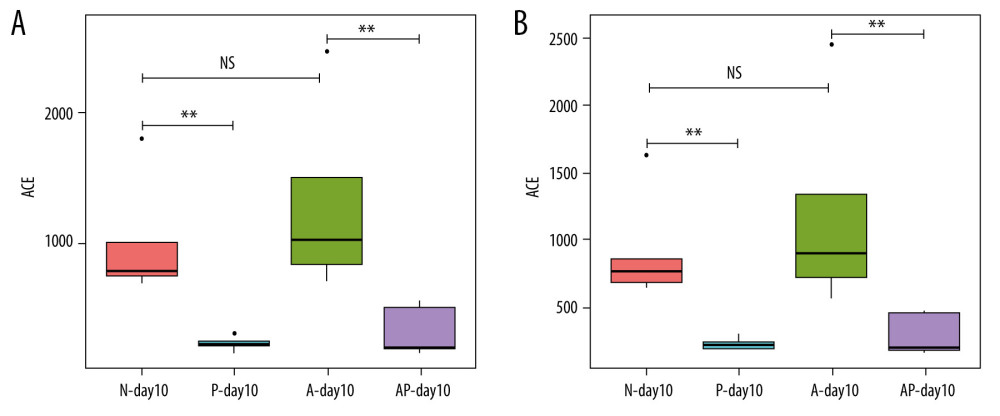 Figure 3. Alpha diversity of gut microflora in the 4 groups at day10. (A) Abundance-based coverage estimator (ACE) index. (B) Chao1 index. N - the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. NS – means no statistical differences. ** P<0.01.
Figure 3. Alpha diversity of gut microflora in the 4 groups at day10. (A) Abundance-based coverage estimator (ACE) index. (B) Chao1 index. N - the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. NS – means no statistical differences. ** P<0.01. 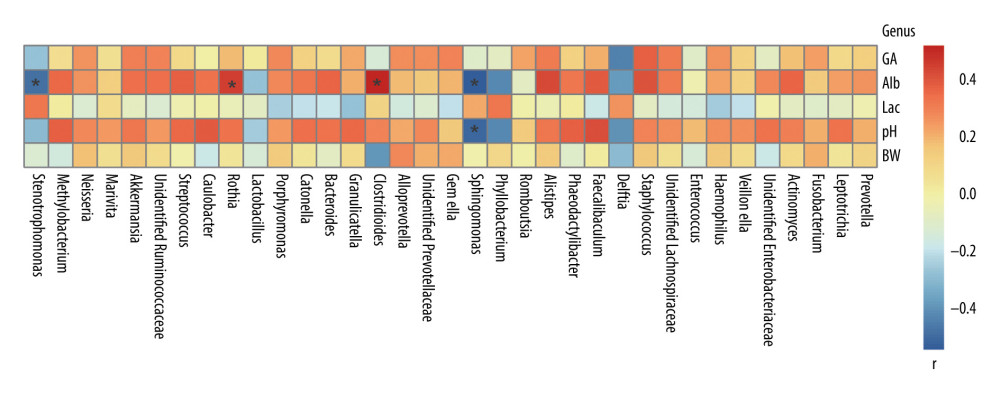 Figure 4. Correlation between gut microbiota and environmental factors of all preterm infants delivered by cesarean section in this study at the genus level. BW – birth weight; GA – gestational age; Alb – blood albumin; Lac – blood lactate; pH – blood pH at birth. r – correlation. * P<0.05.
Figure 4. Correlation between gut microbiota and environmental factors of all preterm infants delivered by cesarean section in this study at the genus level. BW – birth weight; GA – gestational age; Alb – blood albumin; Lac – blood lactate; pH – blood pH at birth. r – correlation. * P<0.05. 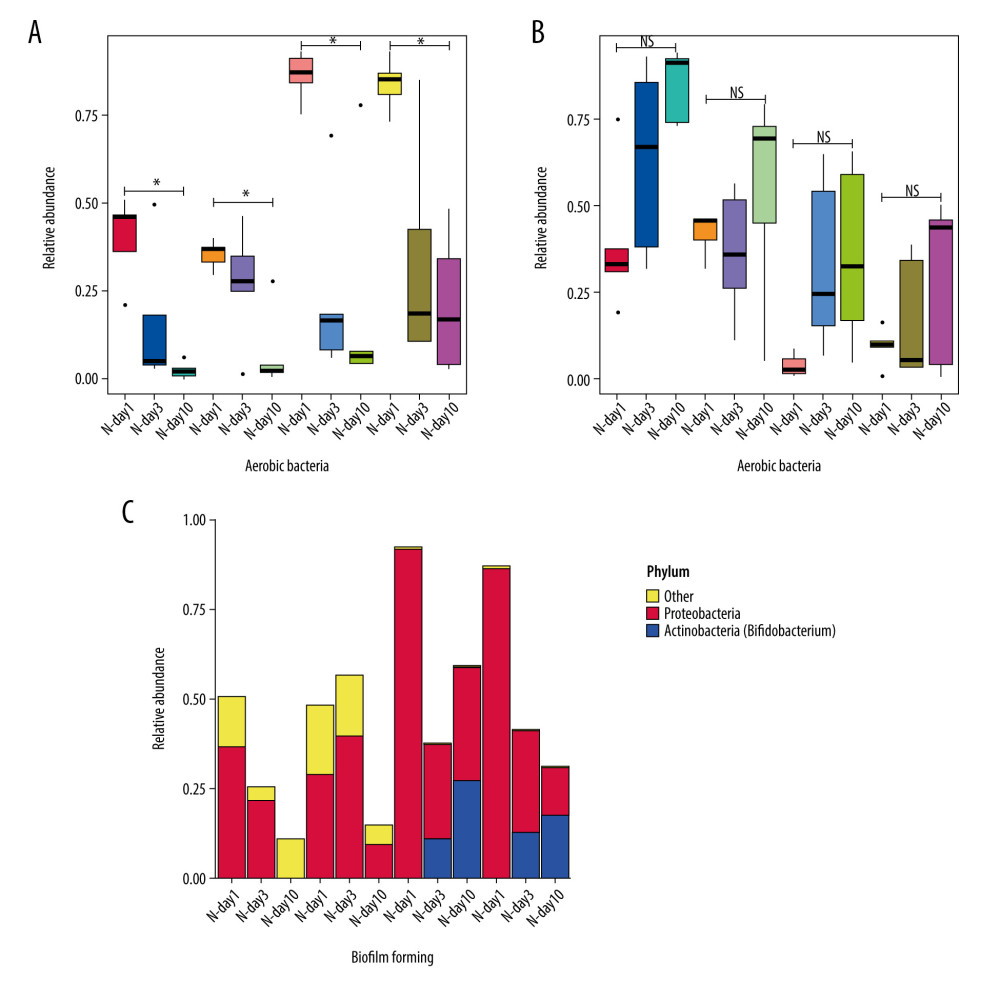 Figure 5. Predicted phenotype of gut microbiota of preterm infants delivered by cesarean section in the 4 groups at 1, 3, and 10 days. (A) Aerobic bacteria relative abundance. (B) Anaerobic bacteria relative abundance. (C) biofilm-forming bacteria relative abundance. N - the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day1 – One day after birth; day3 – Three days after birth; day10 – Ten days after birth. NS – means no significant differences. * P<0.05.
Figure 5. Predicted phenotype of gut microbiota of preterm infants delivered by cesarean section in the 4 groups at 1, 3, and 10 days. (A) Aerobic bacteria relative abundance. (B) Anaerobic bacteria relative abundance. (C) biofilm-forming bacteria relative abundance. N - the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day1 – One day after birth; day3 – Three days after birth; day10 – Ten days after birth. NS – means no significant differences. * P<0.05. 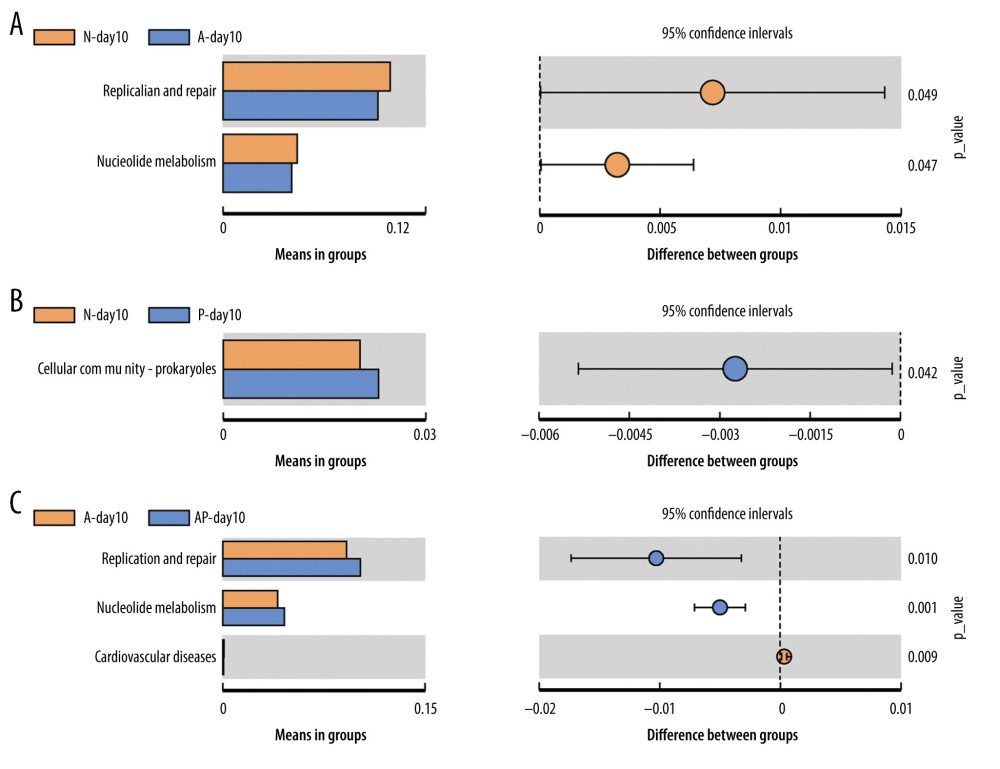 Figure 6. Differences of gut microbiome function of preterm infants delivered by cesarean section in the 4 groups at 10 days. (A) Difference between N-day10 and A-day10 groups. (B) Difference between N-day10 and P-day10 groups. (C) Difference between A-day10 and AP-day10 groups. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. P<0.05 means significant differences.
Figure 6. Differences of gut microbiome function of preterm infants delivered by cesarean section in the 4 groups at 10 days. (A) Difference between N-day10 and A-day10 groups. (B) Difference between N-day10 and P-day10 groups. (C) Difference between A-day10 and AP-day10 groups. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. P<0.05 means significant differences. 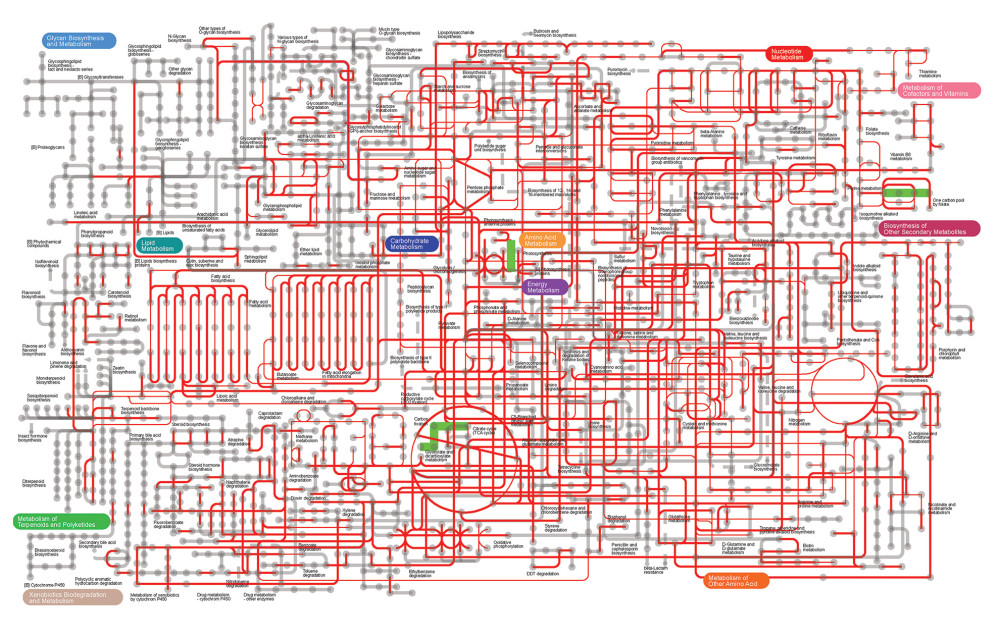 Figure 7. Metabolic pathway prediction between N-day10 and A-day10 groups based on KOs. Red lines are the same metabolic pathway of 2 groups, and Green lines are the metabolic pathway with significant differences of 2 groups (P<0.05). KOs – Kyoto Encyclopedia of Genes and Genomes Orthologous Groups; N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics). Day10 – Ten days after birth.
Figure 7. Metabolic pathway prediction between N-day10 and A-day10 groups based on KOs. Red lines are the same metabolic pathway of 2 groups, and Green lines are the metabolic pathway with significant differences of 2 groups (P<0.05). KOs – Kyoto Encyclopedia of Genes and Genomes Orthologous Groups; N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics). Day10 – Ten days after birth. References
1. Francavilla R, Cristofori F, Tripaldi ME, Indrio F, Intervention for dysbiosis in children born by C-section: Ann Nutr Metab, 2018; 73(3); 33-39
2. Hill CJ, Lynch DB, Murphy K, Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort: Microbiome, 2017; 5(1); 4
3. Arboleya S, Binetti A, Salazar N, Establishment and development of intestinal microbiota in preterm neonates: FEMS Microbiol Ecol, 2012; 79(3); 763-72
4. Dahl C, Stigum H, Valeur J, Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure: Int J Epidemiol, 2018; 47(5); 1658-69
5. Lynch SV, Pedersen O, The human intestinal microbiome in health and disease: New Engl J Med, 2016; 375(24); 2369-79
6. Underwood MA, Sohn K, The microbiota of the extremely preterm infant: Clin Perinatol, 2017; 44(2); 407-27
7. Kuppala VS, Meinzen-Derr J, Morrow AL, Schibler KR, Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants: J Pediatr, 2011; 159(5); 720-25
8. Reyman M, van Houten MA, van Baarle D, Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life: Nat Commun, 2019; 10(1); 1-12
9. Bäckhed F, Roswall J, Peng Y, Dynamics and stabilization of the human gut microbiome during the first year of life: Cell Host Microbe, 2015; 17(5); 690-703
10. Arboleya S, Sánchez B, Milani C, Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics: J Pediatr, 2015; 166(3); 538-44
11. Cantey JB, Pyle AK, Wozniak PS, Early antibiotic exposure and adverse outcomes in preterm, very low birth weight infants: J Pediatr, 2018; 203; 62-67
12. Fouhy F, Guinane CM, Hussey S, High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin: Antimicrob Agents Chemother, 2012; 56(11); 5811-20
13. Robertson RC, Manges AR, Finlay BB, Prendergast AJ, The human microbiome and child growth – first 1000 days and beyond: Trends Microbiol, 2019; 27(2); 131-47
14. Collins A, Weitkamp J-H, Wynn JL, Why are preterm newborns at increased risk of infection?: Arch Dis Child-Fetal, 2018; 103(4); F391-94
15. Pammi M, Cope J, Tarr PI, A systematic review and meta-analysis: Microbiome, 2017; 5(1); 31
16. Budden KF, Gellatly SL, Wood DL, Emerging pathogenic links between microbiota and the gut-lung axis: Nat Rev Microbiol, 2017; 15(1); 55-63
17. Ntranos A, Casaccia P, The microbiome-gut-behavior axis: Crosstalk between the gut microbiome and oligodendrocytes modulates behavioral responses: Neurotherapeutics, 2018; 15(1); 31-35
18. Li YF, Zhu CR, Gong XL, Beneficial effects of probiotic treatment on gut microbiota in very low birth weight infants: Gastroenterol Res Pract, 2019; 2019; 3682836
19. Grzywacz K, Butcher J, Romain G, The impact of probiotics and lactoferrin supplementation on piglet gastrointestinal microbial communities: Biometals, 2019; 32(3); 533-43
20. Akhoundzadeh K, Vakili A, Shadnoush M, Sadeghzadeh J, Effects of the oral ingestion of probiotics on brain damage in a transient model of focal cerebral ischemia in mice: Iran J Med Sci, 2018; 43(1); 32
21. Korpela K, Salonen A, Vepsäläinen O, Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants: Microbiome, 2018; 6(1); 1-11
22. Tang J-n, Zeng Z-g, Wang H-n, An effective method for isolation of DNA from pig faeces and comparison of five different methods: J Microbiol Meth, 2008; 75(3); 432-36
23. Haas BJ, Gevers D, Earl AM, Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons: Genome Res, 2011; 21(3); 494-504
24. Chen C, Yin Q, Wu H, Different effects of premature infant formula and breast milk on intestinal microecological development in premature infants: Front Microbiol, 2020; 10; 3020
25. Kverka M, Zakostelska Z, Klimesova K, Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition: Clin Exp Immunol, 2011; 163(2); 250-59
26. Boente RF, Ferreira LQ, Falcão LS, Detection of resistance genes and susceptibility patterns in Bacteroides and Parabacteroides strains: Anaerobe, 2010; 16(3); 190-94
27. Koizumi A, Maruyama K, Ohki Y: Pediatr Infect Dis J, 2020; 39(6); 546-52
28. Dong Y, Speer CP, Late-onset neonatal sepsis: Recent developments: Arch Dis Child-Fetal, 2015; 100(3); F257-63
29. Fajardo C, Alshaikh B, Harabor A, Prolonged use of antibiotics after birth is associated with increased morbidity in preterm infants with negative cultures: J Matern-Fetal Neo Med, 2019; 32(24); 4060-66
30. Asha N, Tompkins D, Wilcox M: J Clin Microbiol, 2006; 44(8); 2785-91
31. Lau CS, Chamberlain RS: Int J Gen Med, 2016; 9; 27
32. Chernikova DA, Madan JC, Housman ML, The premature infant gut microbiome during the first 6 weeks of life differs based on gestational maturity at birth: Pediatr Res, 2018; 84(1); 71-79
33. Stewart CJ, Nelson A, Scribbins D, Bacterial and fungal viability in the preterm gut: NEC and sepsis: Arch Dis Child-Fetal, 2013; 98(4); F298-303
34. Emami CN, Petrosyan M, Giuliani S, Role of the host defense system and intestinal microbial flora in the pathogenesis of necrotizing enterocolitis: Surg Infect, 2009; 10(5); 407-17
35. Davies D, Understanding biofilm resistance to antibacterial agents: Nat Rev Drug Discov, 2003; 2(2); 114-22
36. Hall CW, Mah T-F, Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria: FEMS Microbiol Rev, 2017; 41(3); 276-301
37. Orrhage K, Nord C, Factors controlling the bacterial colonization of the intestine in breastfed infants: Acta Paediatr, 1999; 88; 47-57
38. Kim Y, Lee JW, Kang S-G: Anaerobe, 2012; 18(5); 539-45
39. Tian H, Zhao L, Zhang Q, The therapeutic effects of magnolia officinalis extraction on an antibiotics-induced intestinal dysbacteriosis in mice: Curr Microbiol, 2020; 77(9); 2413-21
40. Turroni F, Ribbera A, Foroni E, Human gut microbiota and bifidobacteria: From composition to functionality: Antonie Van Leeuwenhoek, 2008; 94(1); 35-50
41. Li J, Lin S, Vanhoutte PM, Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe−/− mice: Circulation, 2016; 133(24); 2434-46
42. Costabile A, Buttarazzi I, Kolida S: PLoS One, 2017; 12(12); e0187964
43. Guo Z, Zhang J, Wang Z, Intestinal microbiota distinguish gout patients from healthy humans: Sci Rep, 2016; 6; 20602
44. Poretsky R, Rodriguez-R LM, Luo C, Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics: PLoS One, 2014; 9(4); e93827
Figures
 Figure 1. The composition of gut microbiome of preterm infants delivered by cesarean section in the 4 groups at 1, 3, and 10 days: Ten most abundant bacteria at the phylum level. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day1 – One day after birth; day3 – Three days after birth; day10 – Ten days after birth.
Figure 1. The composition of gut microbiome of preterm infants delivered by cesarean section in the 4 groups at 1, 3, and 10 days: Ten most abundant bacteria at the phylum level. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day1 – One day after birth; day3 – Three days after birth; day10 – Ten days after birth. Figure 2. Differences of gut microbiome composition of preterm infants delivered by cesarean section in the 4 groups at 10 days. (A) Differences in N-day10, A-day10 and P-day10 group at the genus level. (B) Significance difference Biomarker between the A-day10 and AP-day10 groups. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth.
Figure 2. Differences of gut microbiome composition of preterm infants delivered by cesarean section in the 4 groups at 10 days. (A) Differences in N-day10, A-day10 and P-day10 group at the genus level. (B) Significance difference Biomarker between the A-day10 and AP-day10 groups. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. Figure 3. Alpha diversity of gut microflora in the 4 groups at day10. (A) Abundance-based coverage estimator (ACE) index. (B) Chao1 index. N - the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. NS – means no statistical differences. ** P<0.01.
Figure 3. Alpha diversity of gut microflora in the 4 groups at day10. (A) Abundance-based coverage estimator (ACE) index. (B) Chao1 index. N - the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. NS – means no statistical differences. ** P<0.01. Figure 4. Correlation between gut microbiota and environmental factors of all preterm infants delivered by cesarean section in this study at the genus level. BW – birth weight; GA – gestational age; Alb – blood albumin; Lac – blood lactate; pH – blood pH at birth. r – correlation. * P<0.05.
Figure 4. Correlation between gut microbiota and environmental factors of all preterm infants delivered by cesarean section in this study at the genus level. BW – birth weight; GA – gestational age; Alb – blood albumin; Lac – blood lactate; pH – blood pH at birth. r – correlation. * P<0.05. Figure 5. Predicted phenotype of gut microbiota of preterm infants delivered by cesarean section in the 4 groups at 1, 3, and 10 days. (A) Aerobic bacteria relative abundance. (B) Anaerobic bacteria relative abundance. (C) biofilm-forming bacteria relative abundance. N - the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day1 – One day after birth; day3 – Three days after birth; day10 – Ten days after birth. NS – means no significant differences. * P<0.05.
Figure 5. Predicted phenotype of gut microbiota of preterm infants delivered by cesarean section in the 4 groups at 1, 3, and 10 days. (A) Aerobic bacteria relative abundance. (B) Anaerobic bacteria relative abundance. (C) biofilm-forming bacteria relative abundance. N - the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day1 – One day after birth; day3 – Three days after birth; day10 – Ten days after birth. NS – means no significant differences. * P<0.05. Figure 6. Differences of gut microbiome function of preterm infants delivered by cesarean section in the 4 groups at 10 days. (A) Difference between N-day10 and A-day10 groups. (B) Difference between N-day10 and P-day10 groups. (C) Difference between A-day10 and AP-day10 groups. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. P<0.05 means significant differences.
Figure 6. Differences of gut microbiome function of preterm infants delivered by cesarean section in the 4 groups at 10 days. (A) Difference between N-day10 and A-day10 groups. (B) Difference between N-day10 and P-day10 groups. (C) Difference between A-day10 and AP-day10 groups. N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics); P – the P group (probiotics); AP – the AP group (antibiotics+probiotics). Day10 – Ten days after birth. P<0.05 means significant differences. Figure 7. Metabolic pathway prediction between N-day10 and A-day10 groups based on KOs. Red lines are the same metabolic pathway of 2 groups, and Green lines are the metabolic pathway with significant differences of 2 groups (P<0.05). KOs – Kyoto Encyclopedia of Genes and Genomes Orthologous Groups; N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics). Day10 – Ten days after birth.
Figure 7. Metabolic pathway prediction between N-day10 and A-day10 groups based on KOs. Red lines are the same metabolic pathway of 2 groups, and Green lines are the metabolic pathway with significant differences of 2 groups (P<0.05). KOs – Kyoto Encyclopedia of Genes and Genomes Orthologous Groups; N – the N group (No-probiotics and No-antibiotics); A – the A group (antibiotics). Day10 – Ten days after birth. In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952