06 July 2020: Clinical Research
Microbial Diversity Analysis on Different Surfaces of Dental Restorative Materials via 16S rDNA Sequencing
Yan Tu12ADG, Yuan Wang12ABG, Lingkai Su12CFG, Beibei Shao3F, Zhuhui Duan123ACDE*, Shuli Deng12FGDOI: 10.12659/MSM.923509
Med Sci Monit 2020; 26:e923509
Abstract
BACKGROUND: This study aimed to provide precise material selection guidance for proper clinical restoration and treatment of plaque-related oral diseases, such as dental caries and periodontal diseases.
MATERIAL AND METHODS: Four groups (n=24) of restorative material sheets (n=24) were prepared using 3M Z350 composite resin (ZR), zinc phosphate cement (ZPC), glass-ionomer (GI), and ICON permeable resin (IPR). Six volunteers wore a plaque-collection device equipped with the 4 restorative material sheets for 48 hours. Plaque samples were collected, and Miseq sequencing was applied to obtain template DNA fragments for microbial diversity analysis. The data were analyzed with nonparametric tests.
RESULTS: The microbial diversity on the ZPC surface was significantly lower than that on GI and IPR surfaces. The abundance of Firmicutes and Streptococcus on the ZPC surface was significantly higher than on the surfaces of GI and IPR. In contrast, the abundance of Porphyromonas on the surface of ZPC was significantly lower than that on GI and IPR surfaces. (P<0.05).
CONCLUSIONS: The results of the present study might serve as a basis for material selection under different oral microbial conditions to provide more accurate treatments and restorative procedures in the oral cavity.
Keywords: Dental Materials, Dental Plaque, Sequence Analysis, DNA, Bacteria, Composite Resins, DNA, Ribosomal, Dental Caries, Glass Ionomer Cements, Materials Testing, Resin Cements
Background
There are hundreds of bacteria species in the oral cavity. Oral bacteria-associated conditions are the most general diseases affecting the oral health and even general health [1,2]. It is well established that caries is a chronic infectious disease that has been listed by the World Health Organization (WHO) as one of the 3 major human diseases requiring focal infection prevention and treatment. The term “periodontal disease” refers to a disease status that affects the tooth-supporting structures, and it is the most common cause of tooth loss in the adult population. Moreover, periodontal disease has been confirmed to have a role in cardiovascular diseases, adverse pregnancy outcomes, Alzheimer’s disease, etc. [3–6]. A number of studies have confirmed that in the complex oral environment, dental plaque plays a critical role in the development of caries and periodontitis [7,8]. The treatment of lesions affecting the tooth crown, root caries caused by gingival recession, and wedge-shaped defects mainly rely on the use of a wide variety of dental restorative materials. Dental plaque can form on the surfaces of natural teeth and restorative materials [9,10].
To a large extent, the adhesion, growth, and colonization of bacteria are decided by the nature of the applied restorative materials, which in turn dramatically affects the components and properties of plaque biofilm, and consequently influences the effectiveness and durability of restorations. Currently, a wide variety of restorative materials are used in restorative procedures; however, the selection of these materials depends on patient’s choice and wishes, aesthetics, cost, and the material’s properties and strength. Nonetheless, there is a lack of scientific criteria and clinical guidelines to consider different oral microbial environments in the material selection process. Therefore, it is necessary to investigate microbial diversity on the surfaces of different dental restorative materials to provide theoretical guidance for the selection of these materials.
All the experimental designs for the adhesion and biofilm formation experiments were based on the use of known single or multiple oral bacteria on different restorative materials [11,12]. These designs also reportedly detect demineralization and bacterial invasion of adjacent tooth surfaces and restorative materials [13,14], as well as the width of the gaps between the restorative material and the tooth structure, and the effect of microleakage on secondary or recurrent caries [15–17]. These studies have been
In recent years, the concept of precision medicine has attracted more and more attention. It is a new medical concept; medical models have developed because of rapid advances in genome sequencing technology, the cross-application of biological information, and the use of data technology. With the development of molecular biology, bacteria that could not be cultured and expanded could be detected by molecular biological techniques with a small sample size [20]. 16S rDNA gene-based techniques have already been used to identify oral microbiomes [21–23]. 16S rDNA gene-based techniques have shown changes in the composition and structure of the dental plaque under the disease states [23–25]. Therefore, it is believed that the 16S rDNA gene-based techniques can also be used to investigate microbial diversity on the surface of dental restorative materials
In the present study, a novel lightweight delivery device was designed and constructed to place restorative materials on the coronal surfaces of teeth comfortably and conveniently. The
Material and Methods
RESTORATIVE MATERIALS:
Four different dental restorative materials, including 3M Z350 composite resin (ZR, 3M Filtek™ Z350XT, USA), zinc phosphate cement (ZPC, Stomatology Dental Medicine Material Factory of Wuhan University, China), 3M glass ionomer (GI, 3M ESPE KetacTM Molar Easymix, USA), and ICON permeable resin (IPR, DMG, Germany) were respectively placed in prefabricated sterile molds (3 mm×10 cm) to prepare the cylindrical material block samples. After curing the materials, the blocks were cut into 2–3 mm thick pieces and single-side polished (320–800 mesh) for 30 seconds to 1 minute. Then the samples were subjected to ultraviolet (UV) light for 30 minutes.
PREPARATION OF INTRAORAL APPLIANCES:
Maxillary and mandibular dental casts of the volunteers were prepared according to the complete denture requirements. The dental casts were trimmed and checked for occlusal relationship, maintaining the occlusal relationship when placed on a flat surface. The following design was used for sticking the 4 restorative material sheets on the models: the right upper quadrant, IPR; the left upper quadrant, GI; the left lower quadrant, ZPC; and the right lower quadrant, ZR. Then, alginate impression material was mixed and used to gently fixed a layer of the material sheets on the dental casts; the next step was undertaken after its setting.
The dental lamination machine (vacuum-molding machine) was used to form an invisible retainer. The maxillary and mandibular dental casts were placed, respectively, in the laminated container, and the model was covered with stainless steel sand only enough to expose the dental casts of the teeth, using a vestibular ditch as a standard. Then the lamination machine started to run. The hard-pressed film was softened at 90–150°C for 30–60 seconds, then the film was firmly pressed against the incisors and gingivae of the maxillary or mandibular working model. After the lamination machine was vacuumed, the plywood was automatically disengaged from the maxillary or mandibular dental casts.
Subsequently, the round material sheets were exposed using a special slow-speed handpiece to open a window on the polished sheet and remove the alginate impression material. The edge of the model was trimmed so that it exceeded the gingival margin by 1.8–2.2 mm. Then, the in situ dental plaque model was used. The model was wiped with alcohol and placed under UV light for 30 minutes. Figure 1A–1F presents the schematic representation of the model and its intraoral photographs.
PARTICIPANT SELECTION AND SAMPLE COLLECTION:
This study was approved by the Ethics Committee in the Affiliated Stomatology Hospital of the Zhejiang University Medical College. All the research steps were performed following the relevant guidelines after informed consent was obtained from all the volunteers. Six volunteers were selected, based on the following inclusion criteria: healthy adults with no systemic disease, no use of antibiotics or fluoride compounds within 6 months, no other bacterial or fungal infection in other sites, the presence of at least 24 teeth in the oral cavity, and the absence of caries or periodontal disease.
Each volunteer wore an
DNA EXTRACTION AND PCR AMPLIFICATION:
According to the manufacturer’s protocols, microbial DNA was extracted from 24 samples using the Omega-soil DNA kit (Omega Bio-Tek, Norcross, GA, USA). The V4–V5 region of the bacteria 16S ribosomal RNA gene was amplified by PCR using primers 338F 5′-barcode- ACTCCTACGGGAGGCAGCAG-3′ and 806R 5′-GGACTACHVGGGTWTCTAAT-3′ (95°C for 3 minutes, 95°C for 30 seconds with 27 cycles, 55°C for 30 seconds, and 72°C for 45 seconds and a final extension at 72°C for 10 minutes). PCR reactions were performed in triplicate with a 20 μL mixture containing 4 μL of 5×FastPfu Buffer (TransGen, China), 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu polymerase (TransGen, China), and 10 ng of the template DNA [26].
ILLUMINA MISEQ SEQUENCING:
The amplicons were extracted from 2% agarose gels (Biowest Agarose, Biowest, Spain) and purified by the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor™-ST (Promega, USA). The purified amplicons were integrated into equimolar and paired end sequenced (2×250) on an Illumina MiSeq platform (TruSeq™ DNA Sample Prep Kit, Illumina, USA) conforms to the standard protocols. The raw readings were stored into the NCBI Sequence Read Archive (SRA) database [27].
PROCESSING OF SEQUENCING DATA:
Raw fastq files were decomposed and quality-filtered using QIIME (Version 1.17). The criteria are as follows: 1) the 300 bp readings were truncated at any site receiving an average quality score of <20 over a 50 bp sliding window, discarding the truncated reads that were shorter than 50 bp. 2) Exact barcode matching, 2 nucleotide do not match in primer matching, readings containing indeterminate characters were removed. 3) Only sequences with overlapping length greater than 10 bp were assembled according to their overlap sequence. Readings that could not be gather together were discarded. Operational taxonomic units (OTUs) were clustered using Usearch (Vsesion 7.0
STATISTICAL ANALYSIS:
The variance homogeneity test and the normality test (SPSS 21.0, IBM, USA) of the operational taxonomic unit (OTU) data were obtained. The results showed the inconsistency and non-normal distribution of the data; the variance was not homogeneous. Therefore, the Kruskal-Wallis test with Benjamini-Hochberg false discovery rate (FDR) was performed between the multiple groups and the Wilcoxon rank-sum tests were used for comparison of 2 groups. Statistical significance was set at 0.05 probability level.
Results
GENERAL SEQUENCING DATA ANALYSIS:
Twenty-four samples were sequenced by Miseq 16S rDNA. The total bacterial load was measured from different dental restorative materials, and 850 378 sequences with an average length of 448.12 bp were obtained. The rarefaction curves (Figure 2) of all the samples already reached a platform at this sequencing depth, indicating that the sequencing was deep enough. The sequencing results were blasted with the Silva database, being located to 12 phyla, 75 genera, and 136 species; there were 5 phyla, 22 genera, and 35 species with a proportion of >1%.
At the phylum and genus level (Figure 3), the bacteria on the surfaces of the materials belonged to 5 phyla including Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Actinobacteria, and 22 genera involving Streptococcus, Neisseria, Haemophilus, Veillonella, Porphyromonas, Fusobacterium, Alloprevotella, Prevotella, Gemella, Lautropia, Rothia, Granulicatella, Prevotella 7, Acinetobacter, Unclassified_o_Lactobacillales, Abiotrophia, and Parvimonas. Then the differences between the groups were analyzed in the proportion of >1% of the species.
SPECIES DIVERSITY ANALYSIS:
Differences in bacterial species between the groups were determined by the diversity index, the results of Shannon and Simpson index measurements and statistical analyses to measure the diversity of species are shown in Figure 4. The Shannon index and Simpson indexes (Figure 4A, 4B, * P≤0.05, ** P≤0.01) both showed that the species diversity of the ZPC group was lower than that of the GI and IPR groups, with a significant difference. This means that the adhesion of bacteria to different materials was different. Many research studies have shown that a reduction in microbial diversity in dental plaque often indicates the occurrence of stomatology diseases, such as caries and periodontal disease [8,24,25]. The microbial diversity in oral plaques will change 6 months before caries happens. Therefore, the decline in microbial diversity on the surface of ZPC also reflects the potential problems of traditional oral restorative materials in clinical applications compared to some new materials.
DIFFERENCES IN BACTERIAL ABUNDANCE BETWEEN THE 4 GROUPS:
The results showed that at the phylum and genus level (Figure 5A, 5B), Firmicutes (P=0.043), Streptococcus (P=0.013), and Porphyromonas (P=0.032) exhibited significant differences between the 4 kinds of restorative materials.
These differences between bacteria species have played a role in oral diseases, which cannot be ignored. The increase in Firmicutes has been confirmed as being closely related to dental caries in many studies [20,25,28,29], Streptococcus are Gram-positive bacteria and are often presented in the supragingival plaque; they are associated with the occurrence of dental caries [30,31]. Porphyromonas species is a Gram-negative bacterium that is often found in the subgingival plaque and is often associated with periodontal disease [8,32,33]. These results indicate that ZPC and ZR had a high abundance of Streptococcus on their surfaces compared to the GI and IPR. In addition, GI, IPR, and ZR exhibited a high abundance of Porphyromonas compared to ZPC. Therefore, the differences in the type and abundance of bacteria adhering to the surface of the materials have a significant role in helping the selection and application of clinical restorative materials.
TWO-BY-TWO DIFFERENCES IN BACTERIAL ABUNDANCE:
After discovering the different adhesion patterns of bacteria between the 4 restorative materials, the Wilcoxon nonparametric test was applied for the 2-by-2 comparison of data. Figure 6A–6C presents the results (* represents P≤0.05). At the phylum level, Firmicutes on the IPR surface was lower than that on the ZPC. At the genus level, Streptococcus on the ZPC surface was more numerous than that on the GI and IPR surfaces, Porphyromonas on the ZPC surface was less numerous than that on the GI and IPR surfaces. There was no statistically significant difference between the ZR group and other groups at the phylum level and genus level.
Comparisons between the 4 groups were similar to those of the IPR-ZPC group at phylum and genus levels, indicating that the distinction between the 2 groups was very noticeable, which directly affected the analysis of the results in multiple groups. These conclusions can be seen from the PLS-DA diagram (Figure 7). PLS-DA analysis is a method of testing the similarities and differences between groups. In Figure 7, at the OTU level, IPR and GI sample scatters show the extreme distance from the ZPC samples. However, the ZR and other group samples are close to each other. This is also the reason why ZR exhibited no significant difference from the other groups.
Discussion
At present, many novel restorative materials have antimicrobial effects, including effects on caries-related and periodontitis-related microorganisms [34–37], mainly through the inhibition of the formation of bacterial biofilms and inhibition of the destruction of tooth structure by acid-producing metabolic pathways [38,39]. However, the application of clinical restorative materials is mostly dependent on the cost, aesthetics, the properties of the materials, and patients’ choice and economic conditions, rather than on the individual differences and the effects on micro-ecological diversity in the oral cavity [40,41].
The traditional culture-based methods severely limit the in-depth analysis of the samples. In contrast, Miseq sequencing can provide numerous readings of many samples in a single run, enabling the analysis of several samples at one time. Previous single-bacterial studies and
The oral microecology is basically in a dynamic equilibrium state. Two studies reported that the difference in setting position did not affect the formation of plaque biofilm [7,42]. Therefore, the location of the material does not affect the experimental results. Accordingly, the 4 materials were placed in 4 different quadrants in the oral cavity of each volunteer. To circumvent the differences in bacterial counts due to surface roughness, the same grinding and polishing procedures were carried out on the surfaces of the material sheets to be tested because the surface roughness is positively correlated with the weight of the plaque [11]. To prevent excessive plaque accumulation on the surfaces of the test samples, restorative materials with smooth surfaces were used to inhibit bacterial growth. This is also the reason for the emergence of nano-filled materials that are now available on the market [43], which can be used to provide dental restorations with a smoother surface.
The properties of the restorative materials have been reported to determine the outcomes of bacterial adhesion to a great extent [44–47]. Based on the results of this study (Figure 6), the materials were classified into 3 groups in terms of the ability of different bacterial species to adhere to the materials as follows: the
Interestingly, this study involved the use of a fluoride-containing restorative material, i.e., GI. Previous studies showed that GI has an anti-streptococcal effect that could influence the plaque formation in the early stage [50,51], coincided with the result of the current study, in which the
In general, the rarefaction curve indicated (Figure 2) that the sample size in this study reached the required sequencing depth; therefore, the microbial diversity on the surface of the material obtained in the present study is valid. There were significant differences in the types and abundance of bacteria adhering to the material surfaces in the present study. Firmicutes and
However, Miseq sequencing was not useful at species levels; therefore, the differences between caries- and periodontal disease-related bacteria, such as
Finally, this
Conclusions
The results of the present study can serve as a basis for selecting materials under different oral microbial conditions to optimize oral treatments and restorative procedures.
Figures
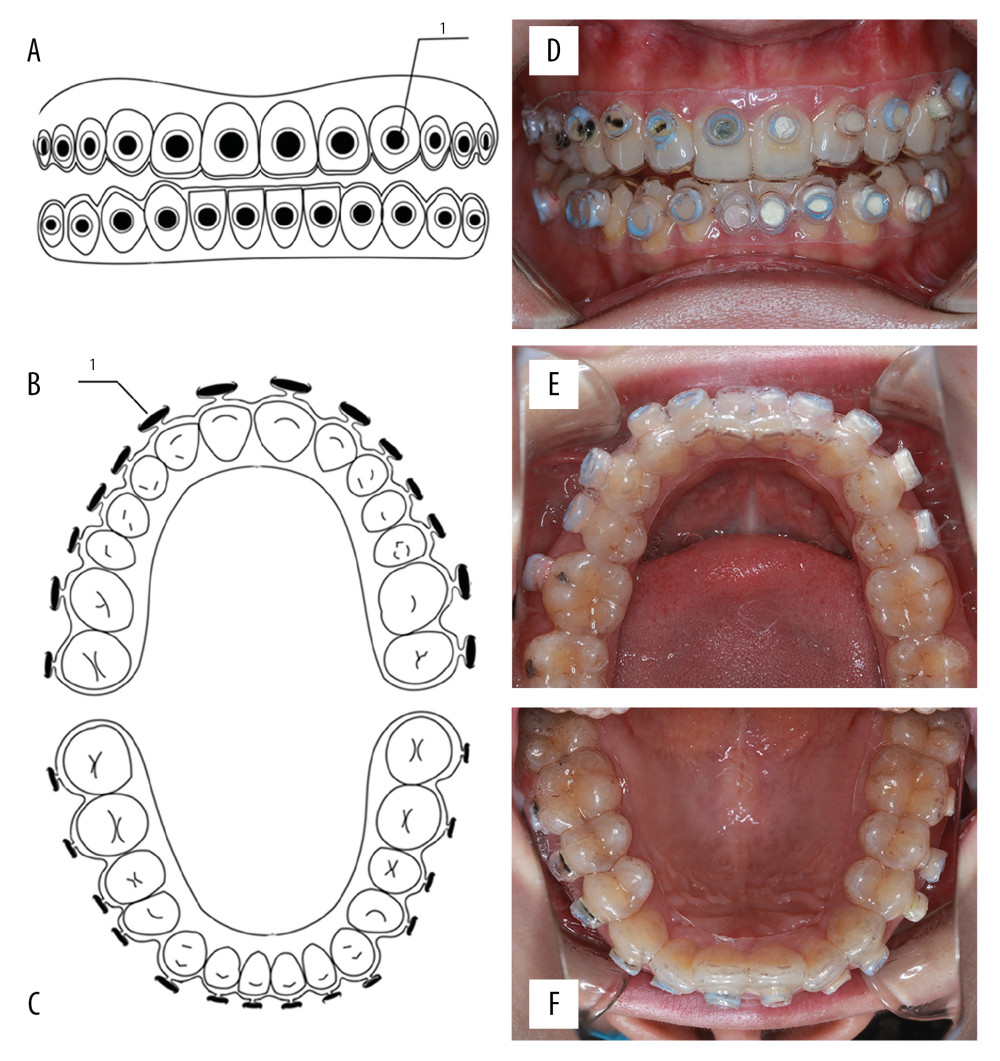 Figure 1. Schematic of the model and intraoral photographs. (A) Front view of the model. (B) Top view of the model (maxillary). (C) Top view of the model (mandibular). (D) Coronal plane. (E) Transverse plane (maxillary). (F) Transverse plane (mandibular). 1 refers to material sheet.
Figure 1. Schematic of the model and intraoral photographs. (A) Front view of the model. (B) Top view of the model (maxillary). (C) Top view of the model (mandibular). (D) Coronal plane. (E) Transverse plane (maxillary). (F) Transverse plane (mandibular). 1 refers to material sheet. 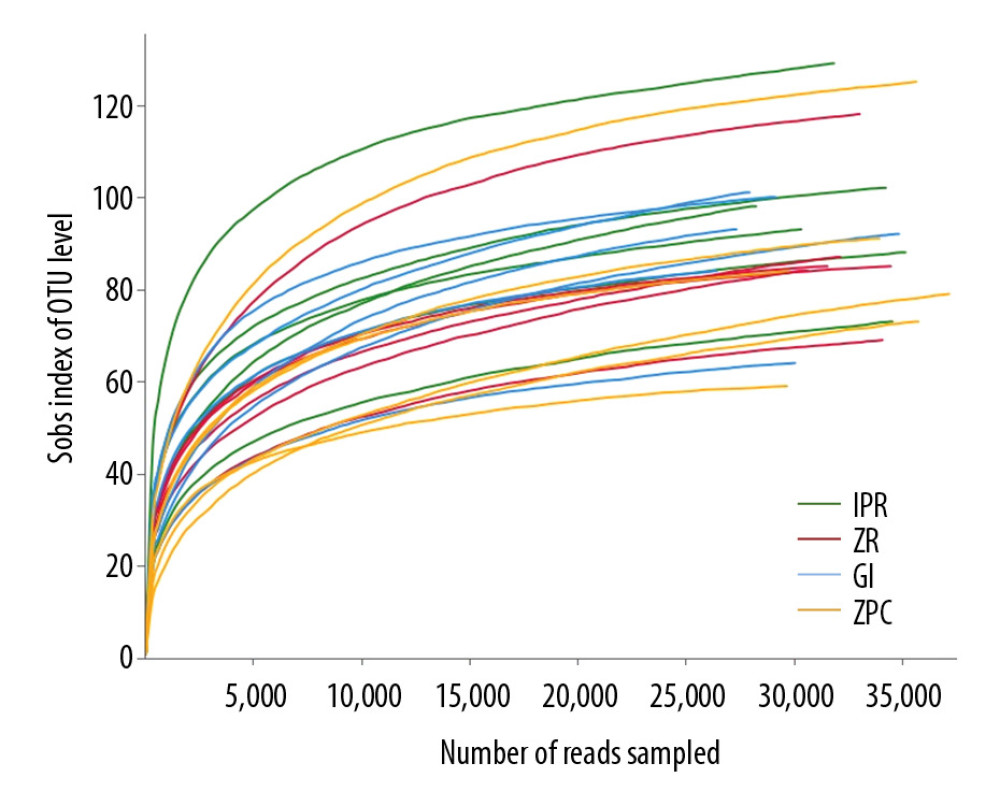 Figure 2. Rarefaction curves. It was used to calculate richness of the total bacterial communities. Vertical axis displays the number of OTUs that would be anticipated to be found after sampling the number of sequences displayed on the horizontal axis. OTUs – operational taxonomic units.
Figure 2. Rarefaction curves. It was used to calculate richness of the total bacterial communities. Vertical axis displays the number of OTUs that would be anticipated to be found after sampling the number of sequences displayed on the horizontal axis. OTUs – operational taxonomic units. 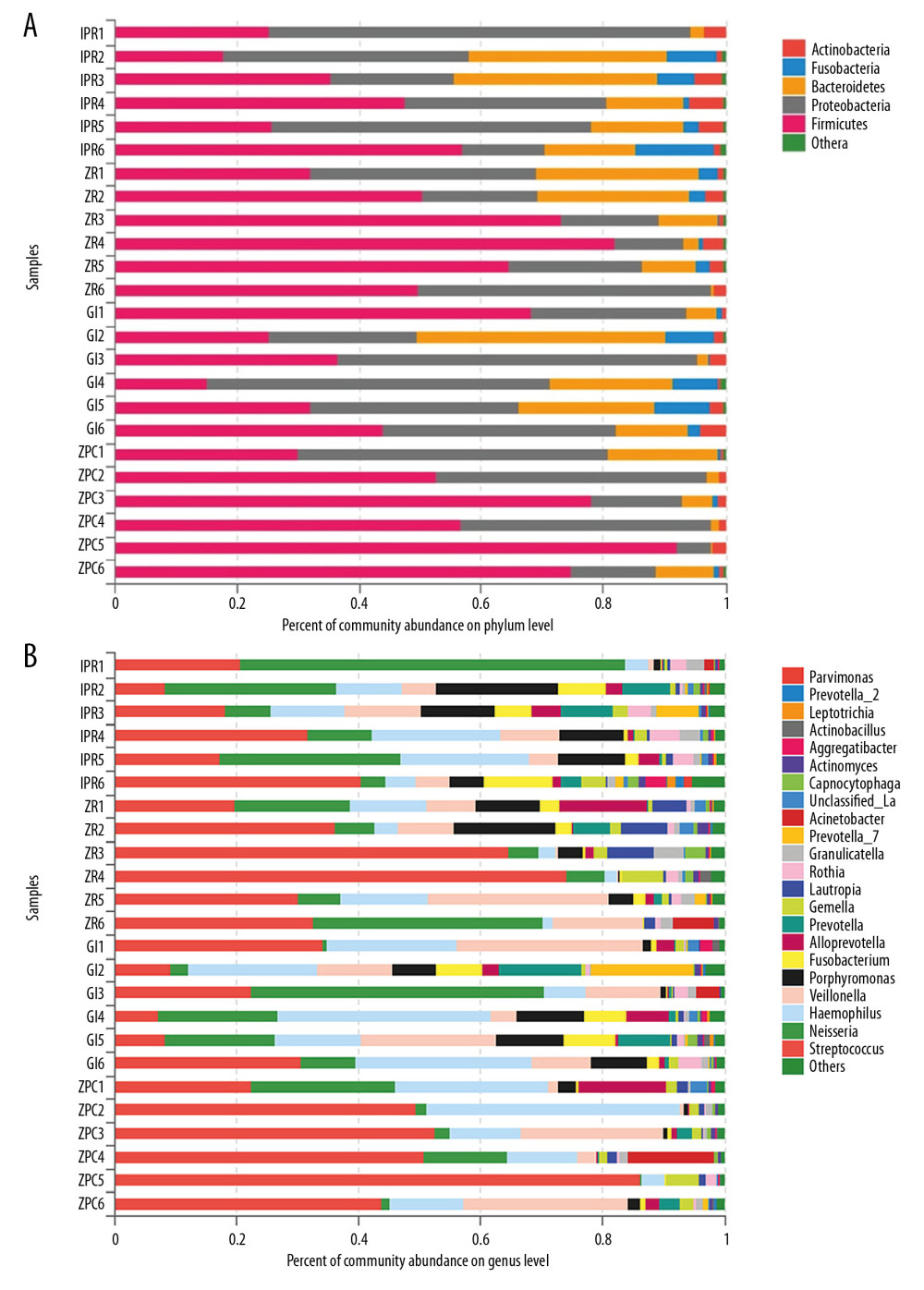 Figure 3. Community barplot at phylum level (A) and genus level (B). The abscissa represents the percent of community abundance on the surface of the materials. The ordinate represents the sample names. Different colors correspond to the different types of phylum and genus on the right side, it can be seen that the bacterial species and abundance on the surface of 4 materials are different. Unclassifed_La refers to unclassified Lactobacillalles.
Figure 3. Community barplot at phylum level (A) and genus level (B). The abscissa represents the percent of community abundance on the surface of the materials. The ordinate represents the sample names. Different colors correspond to the different types of phylum and genus on the right side, it can be seen that the bacterial species and abundance on the surface of 4 materials are different. Unclassifed_La refers to unclassified Lactobacillalles. 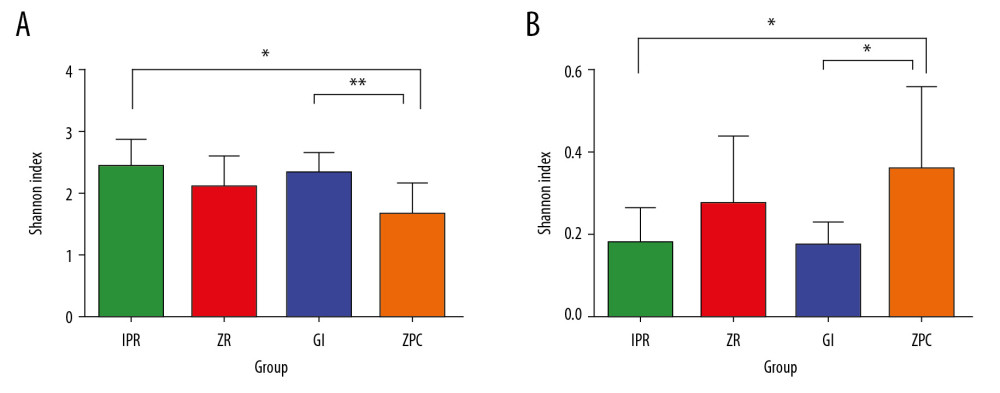 Figure 4. Shannon index (A) and Simpson index (B). These indices were used to estimate microbial diversity between 2 groups. The Shannon index larger, the diversity of the community is higher. In contrast, the Simpson index larger, the diversity of the community is lower. The Shannon index (A) and Simpson index (B) both showed that the species diversity of ZPC group was lower than that of GI and IPR group, and there was a significant difference. * P≤0.05, ** P≤0.01. ZPC – zinc phosphate cement; GI – glass-ionomer; IPR – ICON permeable resin.
Figure 4. Shannon index (A) and Simpson index (B). These indices were used to estimate microbial diversity between 2 groups. The Shannon index larger, the diversity of the community is higher. In contrast, the Simpson index larger, the diversity of the community is lower. The Shannon index (A) and Simpson index (B) both showed that the species diversity of ZPC group was lower than that of GI and IPR group, and there was a significant difference. * P≤0.05, ** P≤0.01. ZPC – zinc phosphate cement; GI – glass-ionomer; IPR – ICON permeable resin. 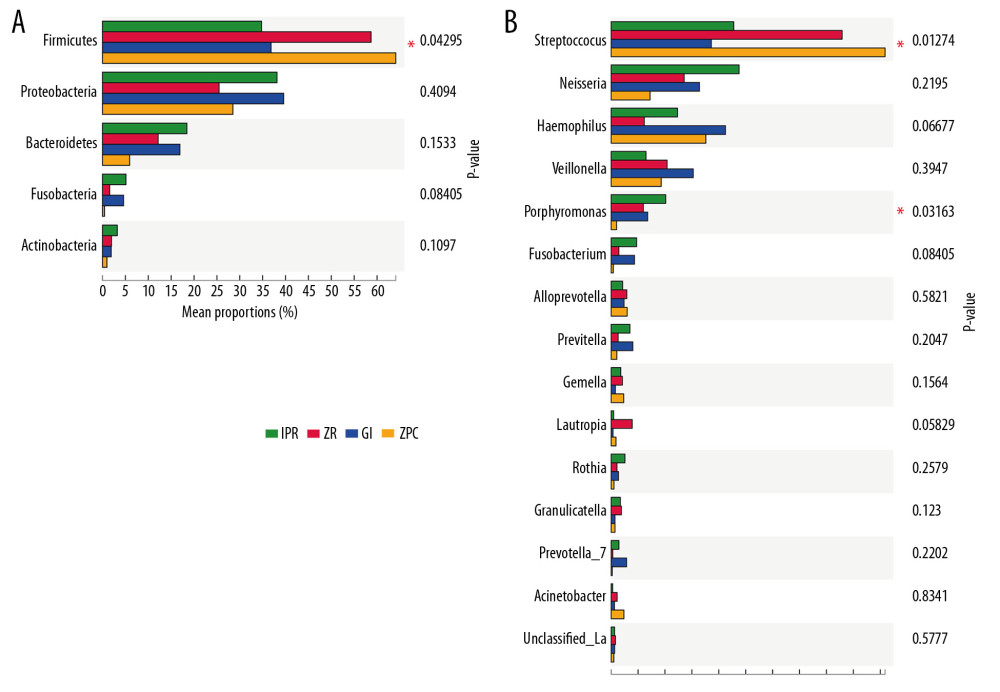 Figure 5. Different taxon analysis among 4 groups at phylum level (A) and genus (B) level. (A) Firmicutes (P=0.043 <0.05) have significant differences at phylum level among 4 groups. (B) Streptococcus (P=0.013 <0.05) and Porphyromonas (P=0.032 <0.05) have significant differences at genus level among 4 groups. The abscissa represents the proportion of bacteria at genus level on the surface of the materials. The left ordinate represents the bacterial species. The right ordinate represents the P value. Unclassifed_La refers to unclassified Lactobacillalles, * represents P≤0.05.
Figure 5. Different taxon analysis among 4 groups at phylum level (A) and genus (B) level. (A) Firmicutes (P=0.043 <0.05) have significant differences at phylum level among 4 groups. (B) Streptococcus (P=0.013 <0.05) and Porphyromonas (P=0.032 <0.05) have significant differences at genus level among 4 groups. The abscissa represents the proportion of bacteria at genus level on the surface of the materials. The left ordinate represents the bacterial species. The right ordinate represents the P value. Unclassifed_La refers to unclassified Lactobacillalles, * represents P≤0.05. 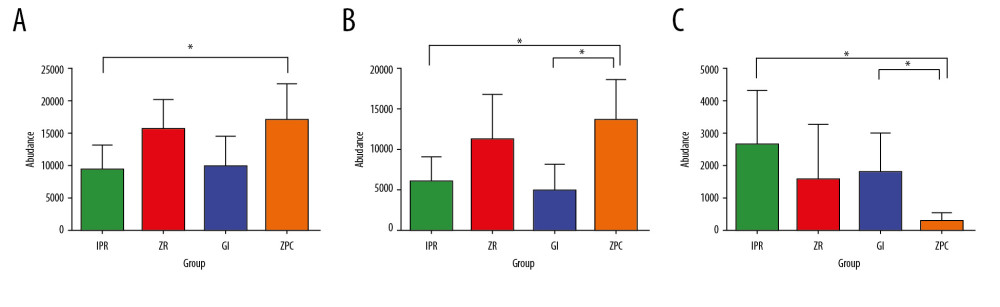 Figure 6. Differential species analysis between 2 groups at phylum level (A) and genus level (B, C). (A) Firmicutes on the IPR surface is lower than ZPC at phylum level. (B) Streptococcus on the ZPC surface is higher than GI and IPR at genus level. (C) Porphyromonas on the ZPC surface is lower than GI and IPR at genus level. * Represents P≤0.05. ZPC – zinc phosphate cement; GI – glass-ionomer; IPR – ICON permeable resin.
Figure 6. Differential species analysis between 2 groups at phylum level (A) and genus level (B, C). (A) Firmicutes on the IPR surface is lower than ZPC at phylum level. (B) Streptococcus on the ZPC surface is higher than GI and IPR at genus level. (C) Porphyromonas on the ZPC surface is lower than GI and IPR at genus level. * Represents P≤0.05. ZPC – zinc phosphate cement; GI – glass-ionomer; IPR – ICON permeable resin. 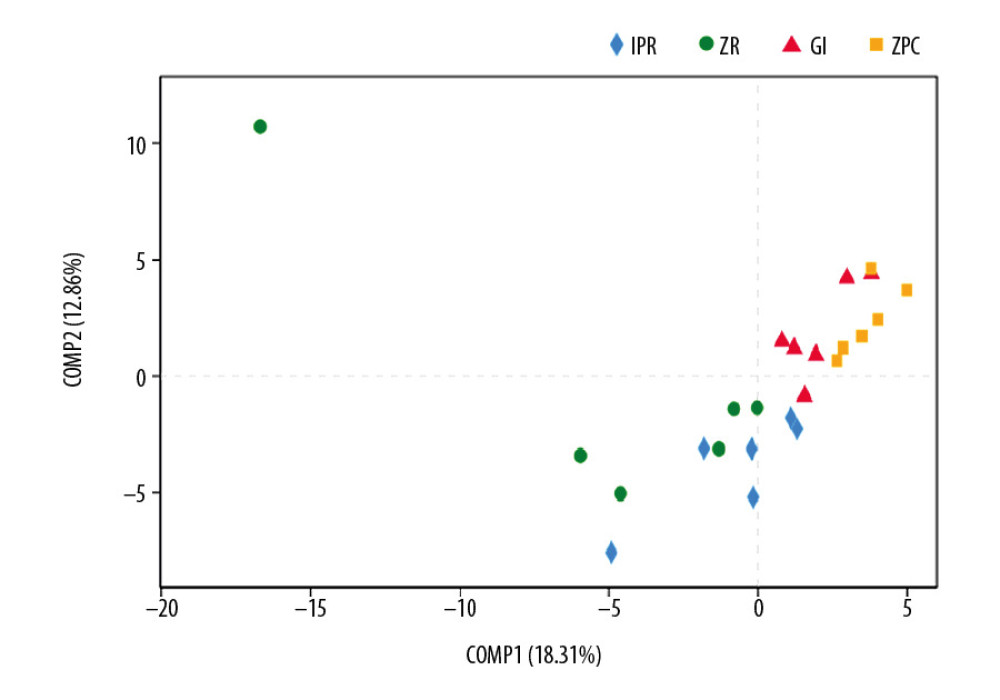 Figure 7. PLS-DA analysis. It is a method of testing the similarity and differences between groups. At the OTU level, IPR and GI sample scatters show the far distance from the ZPC samples. Therefore, there are statistical differences among these experimental groups. OTU – operational taxonomic unit; IPR – ICON permeable resin; GI – glass-ionomer; ZPC – zinc phosphate cement.
Figure 7. PLS-DA analysis. It is a method of testing the similarity and differences between groups. At the OTU level, IPR and GI sample scatters show the far distance from the ZPC samples. Therefore, there are statistical differences among these experimental groups. OTU – operational taxonomic unit; IPR – ICON permeable resin; GI – glass-ionomer; ZPC – zinc phosphate cement. References
1. Sampaio-Maia B, Caldas IM, Pereira ML, The oral microbiome in health and its implication in oral and systemic diseases: Adv Appl Microbiol, 2016; 97; 171-210
2. Krishnan K, Chen T, Paster BJ, A practical guide to the oral microbiome and its relation to health and disease: Oral Dis, 2017; 23(3); 276-86
3. Yamashita Y, Takeshita T, The oral microbiome and human health: J Oral Sci, 2017; 59(2); 201-6
4. Ahmed U, Tanwir F, Association of periodontal pathogenesis and cardiovascular diseases: A literature review: Oral Health Prev Dent, 2015; 13(1); 21-27
5. Chanomethaporn A, Chayasadom A, Wara-Aswapati N, Association between periodontitis and spontaneous abortion: a case-control study: J Periodontol, 2019; 90(4); 381-90
6. Teixeira FB, Saito MT, Matheus FC, Periodontitis and Alzheimer’s disease: A possible comorbidity between oral chronic inflammatory condition and neuroinflammation: Front Aging Neurosci, 2017; 9; 327
7. Auschill TM, Hellwig E, Sculean A, Impact of the intraoral location on the rate of biofilm growth: Clin Oral Investig, 2004; 8(2); 97-101
8. Corrêa JD, Calderaro DC, Ferreira GA, Subgingival microbiota dysbiosis in systemic lupus erythematosus: association with periodontal status: Microbiome, 2017; 5(1); 34
9. Motevasselian F, Zibafar E, Yassini E: J Dent (Tehran), 2017; 14(6); 337-43
10. Ionescu AC, Cazzaniga G, Ottobelli M: J Dent, 2018; 77; 78-86
11. Eick S, Glockmann E, Brandl B, Pfister W: J Oral Rehabil, 2004; 31(3); 278-85
12. Takatsuka T, Konishi N, Nakabo S: Dent Mater J, 2000; 19(4); 363-72
13. Van de Sande FH, Opdam NJ, Truin GJ: J Dent, 2014; 42(9); 1171-77
14. Sousa RP, Zanin IC, Lima JP: J Dent, 2009; 37(1); 44-51
15. Kuper NK, van de Sande FH, Opdam NJ: J Dent Res, 2015; 94(1); 62-68
16. Masih S, Thomas AM, Koshy G, Joshi JL: J Indian Soc Pedod Prev Dent, 2011; 29(2); 135-39
17. Kuper NK, Montagner AF, van de Sande FH: Caries Res, 2015; 49(5); 557-63
18. Wade WG, The oral microbiome in health and disease: Pharmacol Res, 2013; 69(1); 137-43
19. Aas JA, Paster BJ, Stokes LN, Defining the normal bacterial flora of the oral cavity: J Clin Microbiol, 2005; 43(11); 5721-32
20. Gross EL, Leys EJ, Gasparovich SR, Bacterial 16S sequence analysis of severe caries in young permanent teeth: J Clin Microbiol, 2010; 48(11); 4121-28
21. de Melo F, do Nascimento C, Souza DO, de Albuquerque RF, Identification of oral bacteria on titanium implant surfaces by 16S rDNA sequencing: Clin Oral Implants Res, 2017; 28(6); 697-703
22. Li L, Hsiao WW, Nandakumar R, Analyzing endodontic infections by deep coverage pyrosequencing: J Dent Res, 2010; 89(9); 980-84
23. Shchipkova AY, Nagaraja HN, Kumar PS, Subgingival microbial profiles of smokers with periodontitis: J Dent Res, 2010; 89(11); 1247-53
24. Jiang W, Ling Z, Lin X, Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood: Microb Ecol, 2014; 67; 962-69
25. Hao W, Xu H, Chen X, Changes in dental plaque microbial richness and oral behavioral habits during caries development in young Chinese children: Caries Res, 2015; 49(2); 116-23
26. Villhauer AL, Lynch DJ, Drake DR: J Microbiol Methods, 2017; 139; 205-9
27. Pichler M, Coskun ÖK, Ortega-Arbulú AS, A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform: Microbiologyopen, 2018; 7(6); e00611
28. Jiang S, Gao X, Jin L, Lo E, Salivary microbiome diversity in caries-free and caries-affected children: Int J Mol Sci, 2016; 17(12) pii: E1978
29. Caramelli D, Willmann C, Mata X, Oral health status in historic population: Macroscopic and metagenomic evidence: PLoS One, 2018; 13(5); e0196482
30. Mannaa A, Carlén A, Campus G, Campus G, Supragingival plaque microbial analysis in reflection to caries experience: BMC Oral Health, 2013; 13; 5
31. Sbordone L, Bortolaia C, Oral microbial biofilms and plaque-related diseases: Microbial communities and their role in the shift from oral health to disease: Clinical Oral Investig, 2003; 7(4); 181-88
32. Cao Y, Qiao M, Tian Z, Comparative analyses of subgingival microbiome in chronic periodontitis patients with and without IgA nephropathy by high throughput 16S rRNA sequencing: Cell Physiol Biochem, 2018; 47(2); 774-83
33. Ismail FB, Ismail G, Dumitriu AS, Identification of subgingival periodontal pathogens and association with the severity of periodontitis in patients with chronic kidney diseases: A cross-sectional study: Biomed Res Int, 2015; 2015 370314
34. Bai YM, Mao J, Li DX, Bimodal antibacterial system based on quaternary ammonium silane-coupled core-shell hollow mesoporous silica: Acta Biomater, 2019; 85; 229-40
35. Xiao S, Wang H, Liang K, Novel multifunctional nanocomposite for root caries restorations to inhibit periodontitis-related pathogens: J Dent, 2019; 81; 17-26
36. Zhang N, Zhang K, Xie X, Nanostructured polymeric materials with protein-repellent and anti-caries properties for dental applications: Nanomaterials, 2018; 8(6); 393
37. Yue S, Wu J, Zhang Q, Novel dental adhesive resin with crack self-healing, antimicrobial and remineralization properties: J Dent, 2018; 75; 48-57
38. Zhang K, Baras B, Lynch CD, Developing a new generation of therapeutic dental polymers to Inhibit oral biofilms and protect teeth: Materials (Basel), 2018; 11; 1-17
39. Kitagawa H, Miki-Oka S, Mayanagi G, Inhibitory effect of resin composite containing S-PRG filler on Streptococcus mutans glucose metabolism: J Dent, 2018; 70; 92-96
40. Ferracane JL, Hilton TJ, Stansbury JW, Academy of dental materials guidance-resin composites: part II-technique sensitivity (handling, polymerization, dimensional changes): Dent Mater, 2017; 33(11); 1171-91
41. Melo MAS, Weir MD, Passos VF: Int J Mol Sci, 2018; 19(11) pii: E3443
42. Arweiler NB, Hellwig E, Sculean A: Caries Res, 2004; 38(5); 442-47
43. Melo MA, Guedes SF, Xu HH, Rodrigues LK, Nanotechnology-based restorative materials for dental caries management: Trends Biotechnol, 2013; 31(8); 459-67
44. Moberg M, Brewster J, Nicholson J, Roberts H, Physical property investigation of contemporary glass ionomer and resin-modified glass ionomer restorative materials: Clin Oral Investig, 2019; 23(3); 1295-308
45. Henrich B, Hermann I, Di Giulio M: Advances in Materials Science and Engineering, 2016; 2016; 1-12
46. Foley J, Blackwell A: Caries Res, 2003; 37(6); 416-24
47. Ono M, Nikaido T, Ikeda M, Surface properties of resin composite materials relative to biofilm formation: Dent Mater J, 2007; 26(5); 613-22
48. Farrugia C, Haider J, Camilleri L, Camilleri J, Clinical relevance of antimicrobial testing results for dental restorative materials: J Appl Biomater Funct Mater, 2017; 15(2); 153-61
49. Hosida TY, Delbem ACB, Morais LA, Ion release, antimicrobial and physio-mechanical properties of glass ionomer cement containing micro or nanosized hexametaphosphate, and their effect on enamel demineralization: Clin Oral Investig, 2019; 23(5); 2345-54
50. Pandit S, Kim GR, Lee MH, Jeon JG: J Dent, 2011; 39(11); 780-87
51. Wang SP, Ge Y, Zhou XD, Effect of anti-biofilm glass-ionomer cement on Streptococcus mutans biofilms: Int J Oral Sci, 2016; 8(2); 76-83
52. Chhour KL, Nadkarni MA, Byun R, Molecular analysis of microbial diversity in advanced caries: J Clin Microbiol, 2005; 43(2); 843-49
53. Takeshita T, Nakano Y, Kumagai T, The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status: ISME J, 2009; 3(1); 65-78
54. Haririan H, Andrukhov O, Bertl K, Microbial analysis of subgingival plaque samples compared to that of whole saliva in patients with periodontitis: J Periodontol, 2014; 85(6); 819-28
Figures
 Figure 1. Schematic of the model and intraoral photographs. (A) Front view of the model. (B) Top view of the model (maxillary). (C) Top view of the model (mandibular). (D) Coronal plane. (E) Transverse plane (maxillary). (F) Transverse plane (mandibular). 1 refers to material sheet.
Figure 1. Schematic of the model and intraoral photographs. (A) Front view of the model. (B) Top view of the model (maxillary). (C) Top view of the model (mandibular). (D) Coronal plane. (E) Transverse plane (maxillary). (F) Transverse plane (mandibular). 1 refers to material sheet. Figure 2. Rarefaction curves. It was used to calculate richness of the total bacterial communities. Vertical axis displays the number of OTUs that would be anticipated to be found after sampling the number of sequences displayed on the horizontal axis. OTUs – operational taxonomic units.
Figure 2. Rarefaction curves. It was used to calculate richness of the total bacterial communities. Vertical axis displays the number of OTUs that would be anticipated to be found after sampling the number of sequences displayed on the horizontal axis. OTUs – operational taxonomic units. Figure 3. Community barplot at phylum level (A) and genus level (B). The abscissa represents the percent of community abundance on the surface of the materials. The ordinate represents the sample names. Different colors correspond to the different types of phylum and genus on the right side, it can be seen that the bacterial species and abundance on the surface of 4 materials are different. Unclassifed_La refers to unclassified Lactobacillalles.
Figure 3. Community barplot at phylum level (A) and genus level (B). The abscissa represents the percent of community abundance on the surface of the materials. The ordinate represents the sample names. Different colors correspond to the different types of phylum and genus on the right side, it can be seen that the bacterial species and abundance on the surface of 4 materials are different. Unclassifed_La refers to unclassified Lactobacillalles. Figure 4. Shannon index (A) and Simpson index (B). These indices were used to estimate microbial diversity between 2 groups. The Shannon index larger, the diversity of the community is higher. In contrast, the Simpson index larger, the diversity of the community is lower. The Shannon index (A) and Simpson index (B) both showed that the species diversity of ZPC group was lower than that of GI and IPR group, and there was a significant difference. * P≤0.05, ** P≤0.01. ZPC – zinc phosphate cement; GI – glass-ionomer; IPR – ICON permeable resin.
Figure 4. Shannon index (A) and Simpson index (B). These indices were used to estimate microbial diversity between 2 groups. The Shannon index larger, the diversity of the community is higher. In contrast, the Simpson index larger, the diversity of the community is lower. The Shannon index (A) and Simpson index (B) both showed that the species diversity of ZPC group was lower than that of GI and IPR group, and there was a significant difference. * P≤0.05, ** P≤0.01. ZPC – zinc phosphate cement; GI – glass-ionomer; IPR – ICON permeable resin. Figure 5. Different taxon analysis among 4 groups at phylum level (A) and genus (B) level. (A) Firmicutes (P=0.043 <0.05) have significant differences at phylum level among 4 groups. (B) Streptococcus (P=0.013 <0.05) and Porphyromonas (P=0.032 <0.05) have significant differences at genus level among 4 groups. The abscissa represents the proportion of bacteria at genus level on the surface of the materials. The left ordinate represents the bacterial species. The right ordinate represents the P value. Unclassifed_La refers to unclassified Lactobacillalles, * represents P≤0.05.
Figure 5. Different taxon analysis among 4 groups at phylum level (A) and genus (B) level. (A) Firmicutes (P=0.043 <0.05) have significant differences at phylum level among 4 groups. (B) Streptococcus (P=0.013 <0.05) and Porphyromonas (P=0.032 <0.05) have significant differences at genus level among 4 groups. The abscissa represents the proportion of bacteria at genus level on the surface of the materials. The left ordinate represents the bacterial species. The right ordinate represents the P value. Unclassifed_La refers to unclassified Lactobacillalles, * represents P≤0.05. Figure 6. Differential species analysis between 2 groups at phylum level (A) and genus level (B, C). (A) Firmicutes on the IPR surface is lower than ZPC at phylum level. (B) Streptococcus on the ZPC surface is higher than GI and IPR at genus level. (C) Porphyromonas on the ZPC surface is lower than GI and IPR at genus level. * Represents P≤0.05. ZPC – zinc phosphate cement; GI – glass-ionomer; IPR – ICON permeable resin.
Figure 6. Differential species analysis between 2 groups at phylum level (A) and genus level (B, C). (A) Firmicutes on the IPR surface is lower than ZPC at phylum level. (B) Streptococcus on the ZPC surface is higher than GI and IPR at genus level. (C) Porphyromonas on the ZPC surface is lower than GI and IPR at genus level. * Represents P≤0.05. ZPC – zinc phosphate cement; GI – glass-ionomer; IPR – ICON permeable resin. Figure 7. PLS-DA analysis. It is a method of testing the similarity and differences between groups. At the OTU level, IPR and GI sample scatters show the far distance from the ZPC samples. Therefore, there are statistical differences among these experimental groups. OTU – operational taxonomic unit; IPR – ICON permeable resin; GI – glass-ionomer; ZPC – zinc phosphate cement.
Figure 7. PLS-DA analysis. It is a method of testing the similarity and differences between groups. At the OTU level, IPR and GI sample scatters show the far distance from the ZPC samples. Therefore, there are statistical differences among these experimental groups. OTU – operational taxonomic unit; IPR – ICON permeable resin; GI – glass-ionomer; ZPC – zinc phosphate cement. In Press
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
06 Mar 2024 : Clinical Research
Prevalence and Variation of Medical Comorbidities in Oral Surgery Patients: A Retrospective Study at Jazan ...Med Sci Monit In Press; DOI: 10.12659/MSM.943884
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








