16 January 2022: Database Analysis
Junctional Adhesion Molecule-Like Protein (JAML) Is Correlated with Prognosis and Immune Infiltrates in Lung Adenocarcinoma
Likui Fang1ABCDEF, Wenfeng Yu1BCD, Guocan Yu1CDF, Fangming Zhong1DF, Bo Ye1ABCDEF*DOI: 10.12659/MSM.933503
Med Sci Monit 2022; 28:e933503
Abstract
BACKGROUND: Junctional adhesion molecule-like protein (JAML) is a member of the junctional adhesion molecule family and mediates migration of immune cells, but its function in cancers remains unclear. This study aimed to evaluate the role of JAML in the prognosis and immune infiltrates of lung adenocarcinoma (LUAD).
MATERIAL AND METHODS: JAML expressions in LUAD tissues and normal tissues were compared using The Cancer Genome Atlas (TCGA) database and datasets from the Gene Expression Omnibus (GEO) database. The influence of JAML expression on prognosis was analyzed by Kaplan-Meier curve and Cox regression model. Interactive and functional analyses of JAML were performed by LinkedOmics and GeneMANIA databases. TIMER2.0, TISIDB, and GEPIA2 databases were used to investigate the correlation between JAML expression and immune infiltrates.
RESULTS: JAML expression was decreased in LUAD (P<0.001), and lower JAML expression was associated with worse outcomes of LUAD patients. High JAML expression was the protective factor for overall survival (OS) (HR 0.706, 95% CI 0.500-0.997, P=0.048). Interactive and functional analyses suggested that co-expressed genes with JAML have an obvious link to immune-related pathways. In addition, JAML expression was positively associated with infiltrating levels of CD8+ T cells, CD4+ T cells, B cells, dendritic cells, macrophages, and neutrophils, and had significant correlations with diverse immune marker sets in LUAD.
CONCLUSIONS: JAML expression was significantly correlated with prognosis and immune infiltrates. These preliminary findings suggested JAML could be considered as a potential prognostic biomarker and therapeutic target for LUAD.
Keywords: Adenocarcinoma of Lung, JAML Protein, Human, Biomarkers, Tumor, Cell Adhesion Molecules, Female, Gene Expression Profiling, Humans, Lung Neoplasms, Lymphocytes, Tumor-Infiltrating, Male, tumor microenvironment
Background
Lung cancer is one of the most common malignant tumors and is the leading cause of cancer-related death worldwide [1,2]. Lung adenocarcinoma (LUAD) is the most frequent histological subtype [3]. More than half of cases have advanced or metastatic disease at the time of initial diagnosis and require systematic therapy [4]. Although chemotherapy remains an important component of systemic therapy, targeted therapy such as tyrosine kinase inhibitors (TKIs) has shown a better activity in oncogene-driven disease [5,6]. The recent development of immunotherapy has fundamentally shifted the treatment paradigm of patients without sensitive molecular mutations [7]. Immune checkpoint inhibitors (ICIs) have become the front-line treatment in patients with high PD-L1 expression [8]. However, immunotherapy remains ineffective for most patients, and secondary drug resistance is also a challenging problem [9].
Junctional adhesion molecule-like protein (JAML), also named AMICA1, is a junctional adhesion molecule (JAM) that belong to the immunoglobulin superfamily [10]. JAML can mediate the transendothelial migration (TEM) of immune cells and might be involved in modulating cellular responses during tissue immunity [11,12], suggesting that JAML influences the tumor immune microenvironment. In head and neck squamous cell carcinoma, JAML is one of the genes that play a potential role in tumor evolution mechanisms related to the tumor immune microenvironment [13]. However, the function of JAML in LUAD is poorly investigated. This study explored JAML expression in LUAD and its impact on prognosis based on bioinformatics analysis. The association between JAML expression and the tumor immune microenvironment was investigated using the TIMER2.0, TISIDB, and GEPIA2 databases. The results revealed the significant impact of JAML expression on prognosis and tumor immune microenvironment of LUAD.
Material and Methods
DATA ACQUISITION:
The data of mRNA expression and clinicopathologic characteristics of LUAD patients were obtained from The Cancer Genome Atlas (TCGA) database (
LINKEDOMICS DATABASE ANALYSIS:
The LinkedOmics database is a platform (http://www.linkedomics.org) to access, analyze, and compare cancer multi-omics data within and across tumor types [14]. The differentially expressed genes related to JAML were screened through the LinkFinder module in the database, and the correlation was tested using the Pearson correlation coefficient. The gene set enrichment analyses (GSEA) of Gene Ontology biological process (GO_BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were performed in the LinkInterpreter module.
GENEMANIA DATABASE ANALYSIS:
GeneMANIA is a website (http://genemania.org) for generating hypotheses about gene function, analyzing gene lists, and prioritizing genes for functional assays [15]. Given a query gene list, GeneMANIA finds functionally similar genes using a wealth of genomics and proteomics data. We obtained the protein–protein interaction (PPI) network information of JAML using the online tool GeneMANIA.
TIMER2.0 DATABASE ANALYSIS:
The Tumor Immune Estimation Resource (TIMER) is a public website (http://timer.cistrome.org/) providing comprehensive analysis and visualization functions of tumor-infiltrating immune cells [16]. The correlations of JAML expression with the abundance of 6 types of infiltrating immune cells (CD8+ T cells, CD4+ T cells, B cells, dendritic cells, macrophages, and neutrophils) in LUAD were evaluated in the TIMER2.0 database.
TISIDB ANALYSIS:
The Tumor-Immune System Interaction Database (TISIDB) is a user-friendly web portal (http://cis.hku.hk/TISIDB/index.php) which provides comprehensive investigation of tumor-immune interactions [17]. The association between JAML expression and immune features was analyzed in TISIDB to verify the results from TIMER2.0 database analysis.
GEPIA2 DATABASE ANALYSIS:
The Gene Expression Profiling Interactive Analysis (GEPIA) is an online database which provides key interactive and customizable functions, including differential expression analysis, correlation analysis, survival analysis, similar gene detection, and dimensionality reduction analysis [18]. The associations of JAML expression with multiple markers for immune cells were investigated in the GEPIA2 database.
STATISTICAL ANALYSIS:
The
Results
DECREASED JAML EXPRESSION IN LUAD:
The baseline characteristics of the patients with LUAD are presented in Table 1. The JAML expression was significantly lower in LUAD samples in comparison to normal tissue samples in TCGA database (P<0.001, Figure 1A). The ROC curve showed the potential diagnostic value of JAML expression for LUAD (AUC 0.956, 95% CI 0.937–0.975) (Figure 1B). The analyses of datasets (GSE10799, GSE27262, GSE40791, and GSE118370) from the GEO database verified the decreased expression of JAML in LUAD compared to normal tissues (Figure 2A–2D). Correspondingly, the expression of JAML protein was downregulated in LUAD tissue in comparison to that in normal tissue in the Human Protein Atlas (Supplementary Figure 1).
ASSOCIATIONS BETWEEN JAML EXPRESSION AND CLINICOPATHOLOGIC CHARACTERISTICS:
The associations between JAML expression and clinicopathologic characteristics were analyzed to explore the role of JAML in LUAD progress. The results showed no significant association between JAML expression and age (Figure 3A), with lower expression of JAML in males and smokers (P=0.003 and 0.019, respectively, Figure 3B, 3C). T1 stage had higher expression of JAML than T2 and T3 stage (P=0.001 and 0.042, respectively), but the differential expression was not observed in T4 stage (Figure 4A). In addition, JAML expression decreased in positive N stage (N1, N2, and N3) and advanced pathologic stage (Stage III and IV) compared to N0 and early pathologic stage (Stage I and II) (P=0.021 and 0.006, respectively, Figure 4B, 4D), but JAML expression was not significantly associated with M stage (Figure 4C).
LOW JAML EXPRESSION SHOWING POOR SURVIVAL OUTCOMES:
Survival analyses by Kaplan-Meier method and the log-rank test showed that the patients with low JAML expression were correlated with inferior OS (P=0.001) and PFS (P=0.041) (Figure 5A, 5B). Univariate Cox regression analysis demonstrated that JAML expression was the significant factor for OS (HR 0.618, 95% CI 0.460–0.831) and PFS (HR 0.758, 95% CI 0.581–0.989) (Table 2). Multivariate Cox regression analysis proved that high JAML expression was the independent protective factor for OS (HR 0.706, 95% CI 0.500–0.997, P=0.048) (Figure 6).
INTERACTION ANALYSIS OF JAML IN LUAD:
The LinkFinder module in the LinkedOmics database was used to explore the co-expression pattern of JAML in LUAD. The number of observations in the search dataset was 515. A total of 10 820 genes (red dots) were positively correlated with JAML and 9168 genes (green dots) were negatively correlated (Supplementary Figure 2). The top 50 genes that were positively associated with JAML are presented in Figure 7A and the top 50 genes negatively associated with JAML are presented in Figure 7B. These genes are listed in Supplementary Tables 1 and 2, respectively.
In the LinkInterpreter module, GO term annotation showed that co-expressed genes of JAML were mainly involved in interleukin-4 production, cellular defense response, interferon-gamma production, interleukin-1 production, interleukin-2 production, leukocyte proliferation, T cell activation, adaptive immune response, leukocyte cell-cell adhesion, and immune response-regulation signaling pathway (Figure 7C). KEGG pathway analysis indicated enrichment in autoimmune thyroid disease, Staphylococcus aureus infection, asthma, hematopoietic cell lineage, viral myocarditis, allograft rejection, intestinal immune network for IgA production, cell adhesion molecules, and Th1 and Th2 cell differentiation (Figure 7D).
The results from GeneMANIA database analysis revealed that the functions of JAML and their associated molecules were primarily related to leukocyte and neutrophil migration, cell adhesion molecule binding, neutrophil and leukocyte chemotaxis, and leukocyte cell-cell adhesion (Figure 8).
CORRELATIONS OF JAML EXPRESSION WITH IMMUNE INFILTRATES:
The correlation of JAML expression with immune cell infiltration was explored in the TIMER2.0 database. The result showed that JAML expression was significantly and positively associated with the infiltrating levels of CD8+ T cells, CD4+ T cells, B cells, dendritic cells, macrophages, and neutrophils (Figure 9). In TISIDB, JAML expression was also correlated with abundance of activated CD8+ T cells, activated CD4+ T cells, activated B cells, activated dendritic cells, macrophages, and neutrophils (Figure 10).
To further investigate the relationship between JAML and diverse immune infiltrating cells, the correlations between JAML and immune marker sets of immune cells in LUAD were analyzed in the GEPIA2 database. We found that the expression levels of most marker sets marking different T cells, B cell, regulatory T cell (Treg), tumor-associated macrophage (TAM), monocytes, neutrophils, and dendritic cell (DC) had significant correlations with JAML expression in LUAD (Table 3). Marker genes of T cell exhaustion also had a positive correlation with JAML expression, especially TIM-3.
Discussion
Tumorigenesis involves a succession of complex genetic alterations, and avoiding immune destruction is one of the critical processes [19]. T cell infiltration is widely accepted as a key component of adaptive cancer immune and tumor-infiltrating lymphocytes within the tumor microenvironment and are considered prognostic markers in various cancers [20,21]. JAMs are intercellular adhesion molecules that importantly participate in the transmigration of monocytes, neutrophils, and some T cells through homophilic and heterophilic interactions [22]. Increasing evidence suggests that JAM family members could have potential clinical relevance in cancer development and function as tumor suppressors [23,24]. JAML has been reported as a new JAM family member, and its role in LUAD has not yet been fully elucidated.
This study explored the effect of JAML expression on the progress and prognosis of LUAD on the basis of various public databases. By analyzing the data from TCGA and GEO databases, we found that LUAD samples had lower JAML expression compared to normal tissue samples. Decreased JAML expression was observed in advanced T stage, N stage, and pathologic stage, while the differential expression was not significant in T4 stage and M stage. In addition, the survival analysis revealed that high expression of JAML in LUAD had favorable OS and PFS in comparison to low expression. Cox regression analysis further proved the protective role of high JAML expression in OS. These results suggested that JAML is closely correlated with LUAD tumorigenesis and progression and might act as a tumor suppressor. Two recent bioinformatics studies have also demonstrated that JAML is one of the favorable genes in LUAD, and low JAML expression could increase lung cancer risk [25,26]. However, its role in the tumor immune microenvironment was unknown.
Previous studies indicated that JAML plays an important role in γδ T cell activation and tissue homeostasis [27,28]. JAML was also identified as a crucial component of DC migration and can promote response to DC-based cancer immunotherapy [29]. Similarly, our study demonstrated the significant association between JAML and the tumor immune microenvironment. LinkedOmics database analysis pointed out that most co-expressed genes with JAML had obvious links to immune-related pathways, which agrees with the result of GeneMANIA database analysis. Moreover, JAML expression had a significantly consistent correlation with the infiltration levels of CD8+ T cells, CD4+ T cells, B cells, dendritic cells, macrophages, and neutrophils. The correlation between JAML expression and the marker genes of immune cells also suggests the role of JAML in recruitment and regulation of immune infiltrating cells. First, JAML expression was associated with most markers of T helper cells, which suggested that JAML could potentially regulate T cell functions in LUAD. Second, significant correlations were also found between JAML expression and markers of monocytes. Intriguingly, JAML expression was positively correlated with the expression of Treg and T cell exhaustion markers, which contributed to the formation of an immunosuppressive microenvironment. A recent report demonstrated that high frequency of circulating Tregs was associated with a favorable response to ICIs therapy [30]. In addition, M2 macrophage markers such as VSIG4 and MS4A4A showed strong correlations with JAML expression, while M1 macrophage markers showed weak correlations. These results revealed the potential role of JAML in polarization of TAM. Taken together, our findings suggest that JAML is specifically correlated with immune infiltrating cells and plays a vital role in immune regulation in the LUAD microenvironment, but the interaction between JAML and the immune microenvironment is complicated, and does not just lead to a completely positive or negative effect.
There were several limitations in our study. First, all the data analyzed in our study were obtained from online databases, and data heterogeneity was difficult to avoid. Cox regression analysis showed M stage was not significantly associated with PFS. This discrepancy might be caused by data heterogeneity and limited samples. Second, the role of JAML in regulating immune microenvironment seemed to be complicated, and the details of mechanisms by which JAML regulates T cell functions remain unknown. Finally, our results need to be validated by further studies.
Conclusions
Our preliminary findings showed that JAML was downregulated in LUAD, and low JAML expression was associated with tumor progression and poor prognosis. High JAML expression was an independent protective factor for OS and could be considered as a prognostic biomarker. Because of the significant association between JAML expression and the infiltration of diverse immune cells, JAML might play important roles in regulation of the immune microenvironment in LUAD.
Figures
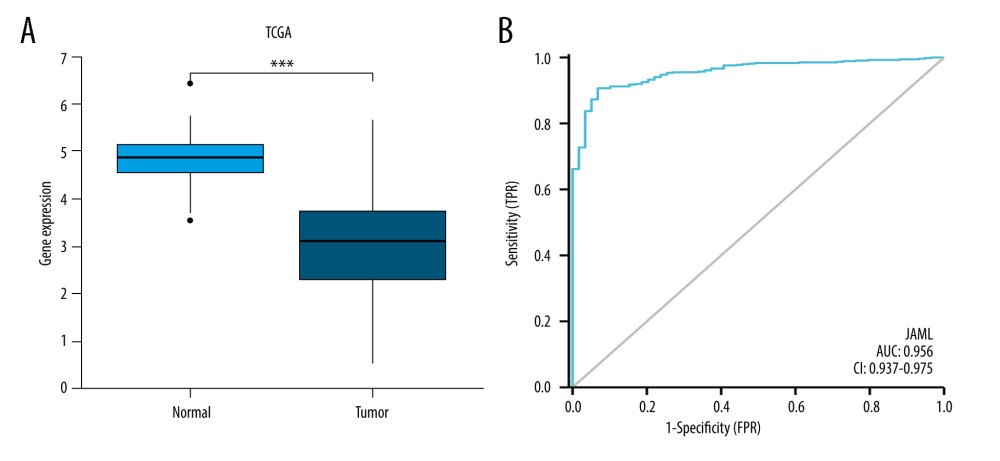 Figure 1. (A) JAML expression was lower in LUAD tissues than in normal tissues in TCGA database. (B) The ROC curve of JAML expression in LUAD. TPR – true positive rate; FPR – false positive rate. *** P<0.001. The figure was created using R statistical software (version 3.6.3).
Figure 1. (A) JAML expression was lower in LUAD tissues than in normal tissues in TCGA database. (B) The ROC curve of JAML expression in LUAD. TPR – true positive rate; FPR – false positive rate. *** P<0.001. The figure was created using R statistical software (version 3.6.3). 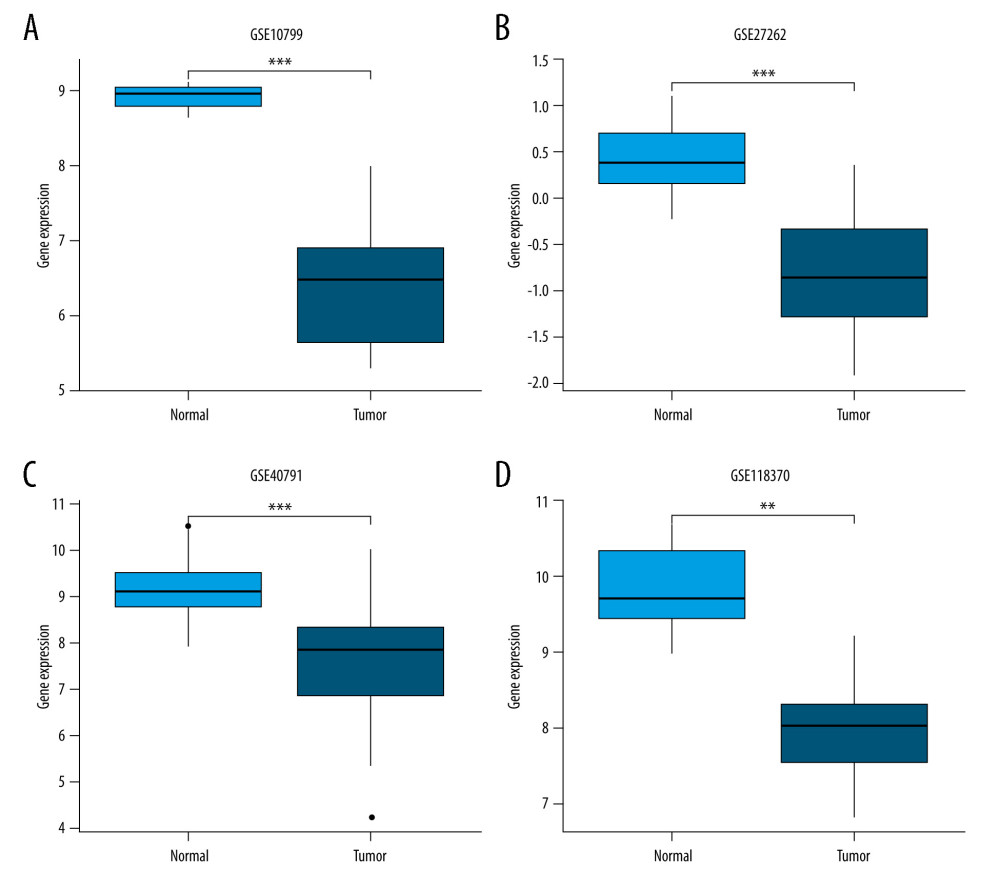 Figure 2. Validation of lower JAML expression in LUAD than that in normal tissues in (A) GSE10799, (B) GSE27262, (C) GSE40791, and (D) GSE118370 datasets. ** P<0.01; *** P<0.001. The figure was created using R statistical software (version 3.6.3).
Figure 2. Validation of lower JAML expression in LUAD than that in normal tissues in (A) GSE10799, (B) GSE27262, (C) GSE40791, and (D) GSE118370 datasets. ** P<0.01; *** P<0.001. The figure was created using R statistical software (version 3.6.3). 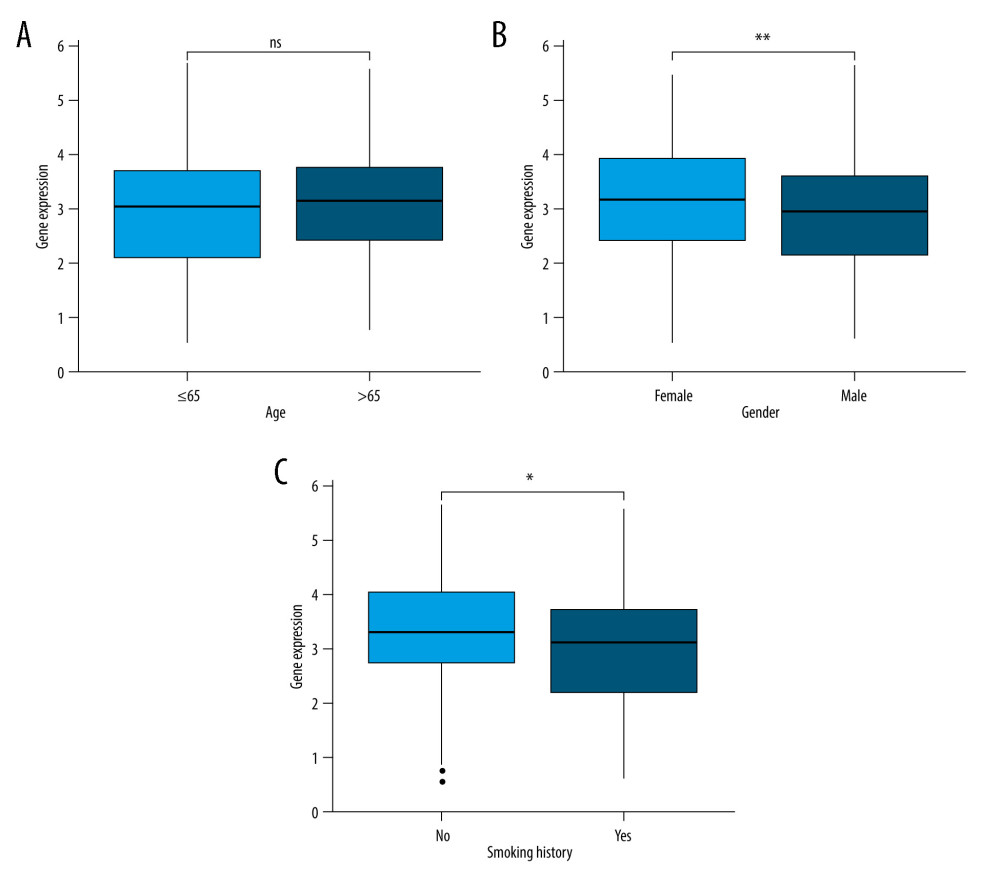 Figure 3. (A) There was no significant difference between JAML expression and age. (B) JAML expression was lower in males than in females. (C) JAML expression was lower in patients with smoking history than in those without smoking history. * P<0.05; ** P<0.01. The figure was created using R statistical software (version 3.6.3).
Figure 3. (A) There was no significant difference between JAML expression and age. (B) JAML expression was lower in males than in females. (C) JAML expression was lower in patients with smoking history than in those without smoking history. * P<0.05; ** P<0.01. The figure was created using R statistical software (version 3.6.3). 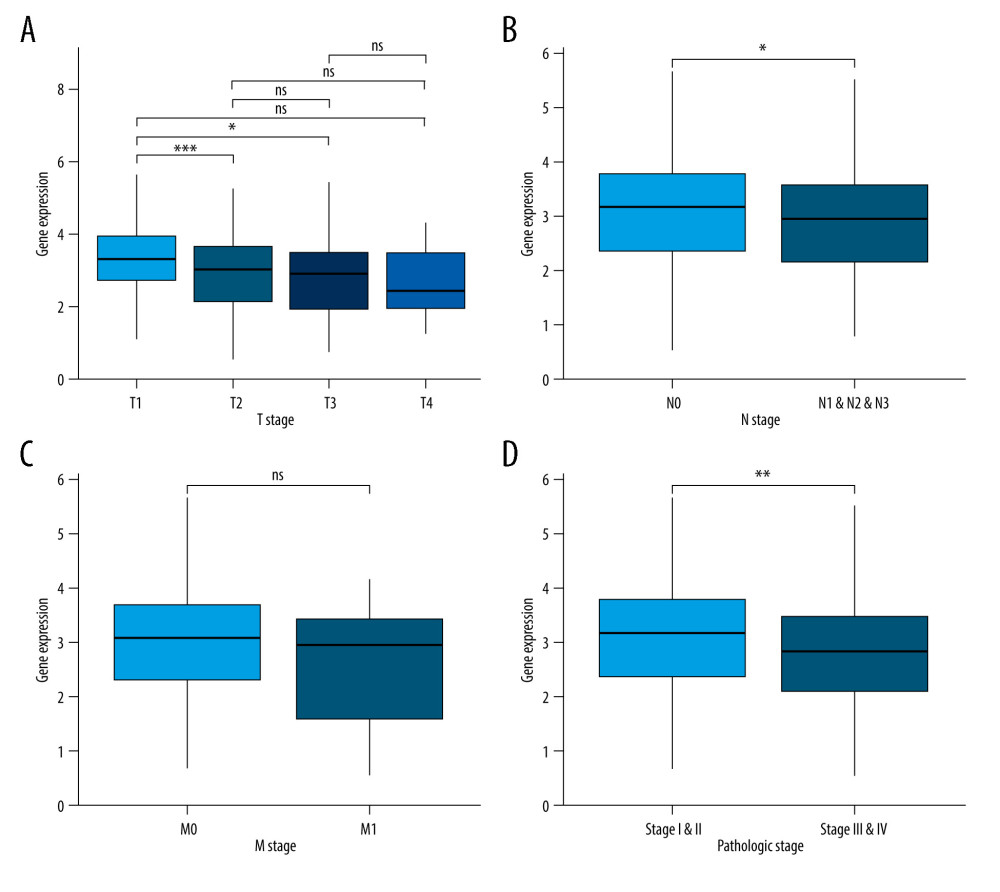 Figure 4. (A) JAML expression was lower in T2 and T3 stage than in T1 stage, but the difference was not observed in T4 stage. (B) Low JAML expression was associated with positive N stage. (C) There was no significant difference between JAML expression and M stage. (D) Low JAML expression was associated with advanced pathologic stage. ns, P≥0.05; * P<0.05; ** P<0.01; *** P<0.001. The figure was created using R statistical software (version 3.6.3).
Figure 4. (A) JAML expression was lower in T2 and T3 stage than in T1 stage, but the difference was not observed in T4 stage. (B) Low JAML expression was associated with positive N stage. (C) There was no significant difference between JAML expression and M stage. (D) Low JAML expression was associated with advanced pathologic stage. ns, P≥0.05; * P<0.05; ** P<0.01; *** P<0.001. The figure was created using R statistical software (version 3.6.3). 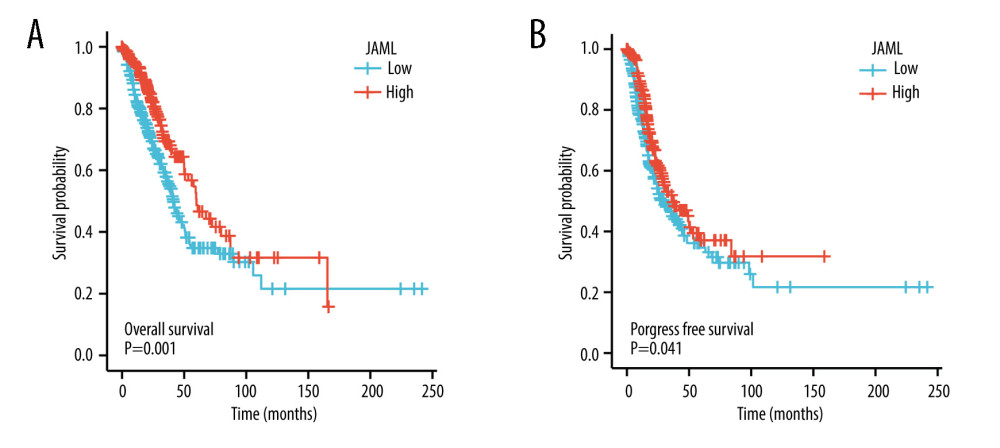 Figure 5. Kaplan-Meier survival curves for (A) overall survival (OS) and (B) progression-free survival (PFS) of the LUAD patients with high and low JAML expression level. The figure was created using R statistical software (version 3.6.3).
Figure 5. Kaplan-Meier survival curves for (A) overall survival (OS) and (B) progression-free survival (PFS) of the LUAD patients with high and low JAML expression level. The figure was created using R statistical software (version 3.6.3). 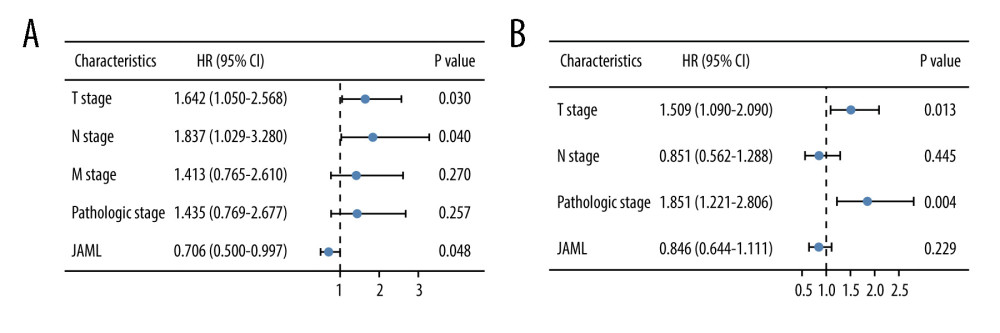 Figure 6. Multivariate Cox regression analysis of JAML expression and clinicopathologic characteristics with (A) overall survival (OS) and (B) progression-free survival (PFS) in LUAD. The figure was created using R statistical software (version 3.6.3).
Figure 6. Multivariate Cox regression analysis of JAML expression and clinicopathologic characteristics with (A) overall survival (OS) and (B) progression-free survival (PFS) in LUAD. The figure was created using R statistical software (version 3.6.3). 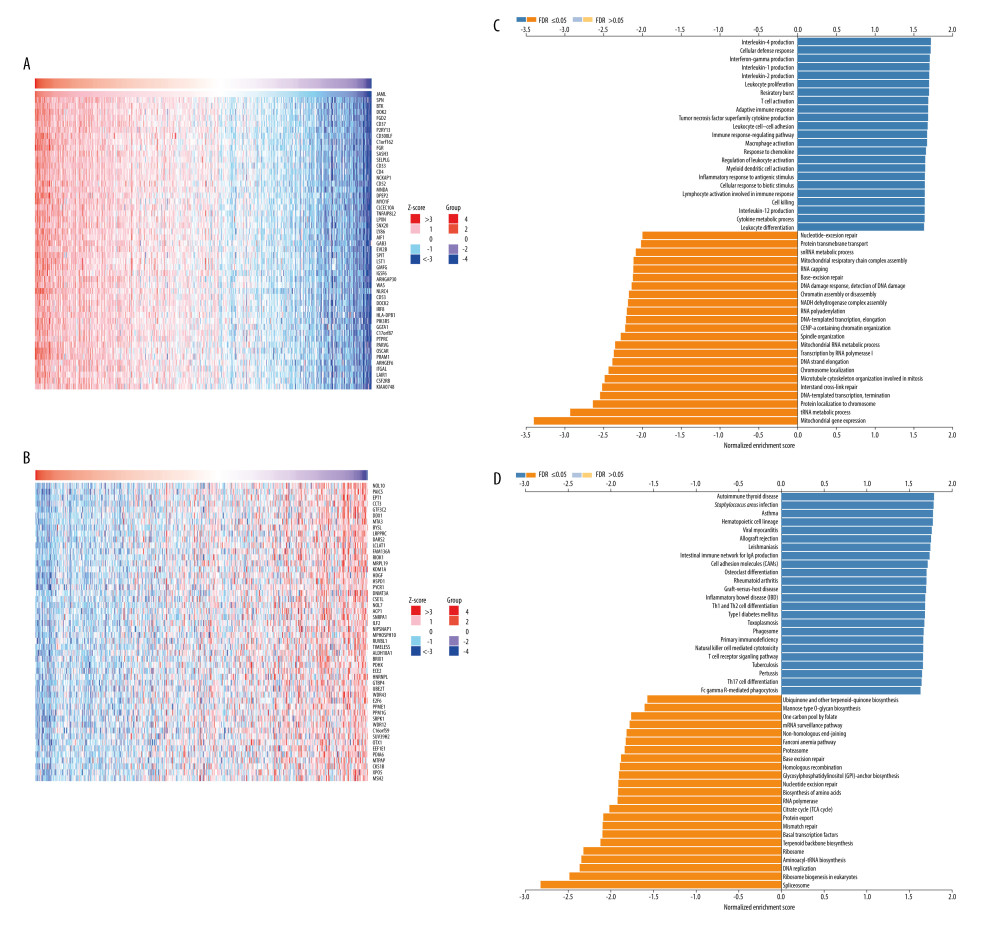 Figure 7. The co-expression genes with JAML in LUAD from the LinkedOmics database. (A, B) Heat maps of top 50 genes positively and negatively correlated with JAML in LUAD, respectively. (C, D) GO annotations and KEGG pathways of JAML in LUAD. The figure was created using the LinkedOmics database (http://www.linkedomics.org).
Figure 7. The co-expression genes with JAML in LUAD from the LinkedOmics database. (A, B) Heat maps of top 50 genes positively and negatively correlated with JAML in LUAD, respectively. (C, D) GO annotations and KEGG pathways of JAML in LUAD. The figure was created using the LinkedOmics database (http://www.linkedomics.org). 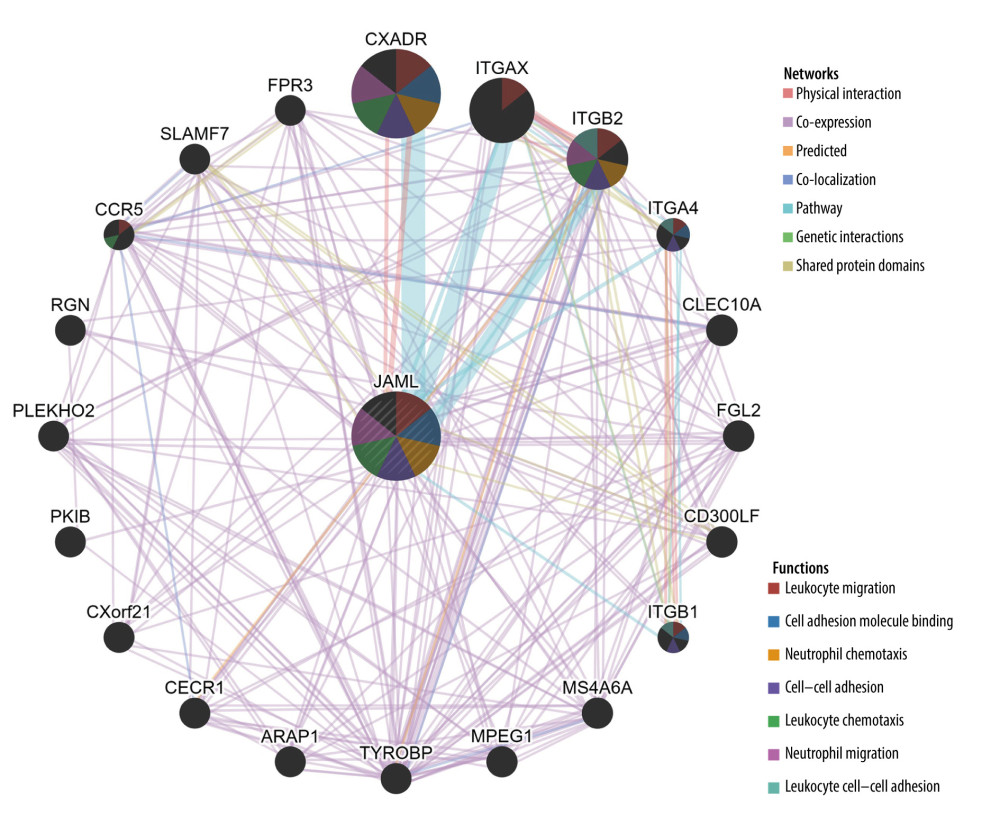 Figure 8. Protein–protein interaction network of JAML. The figure was created using the GeneMANIA database (http://genemania.org).
Figure 8. Protein–protein interaction network of JAML. The figure was created using the GeneMANIA database (http://genemania.org). 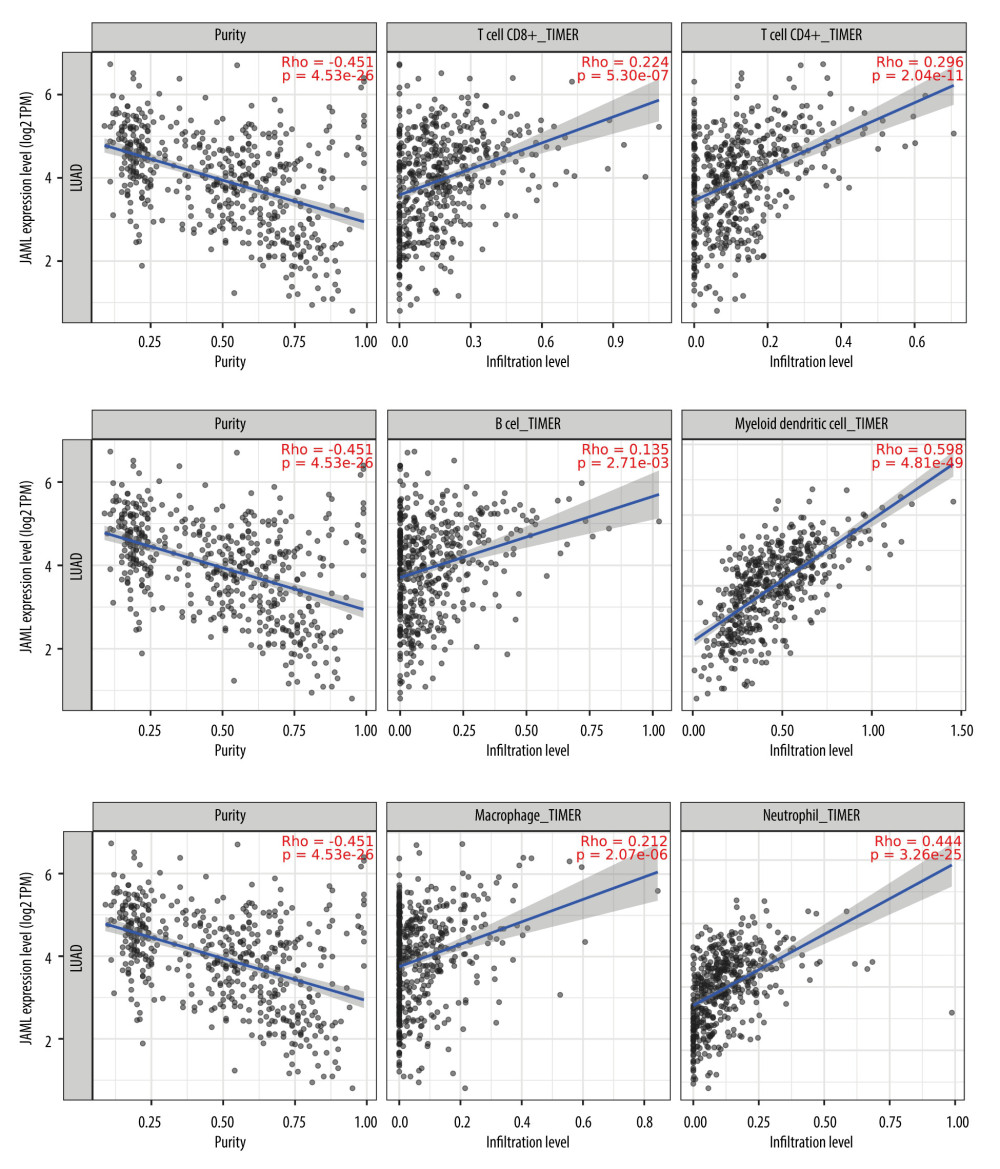 Figure 9. Correlation of JAML expression with immune infiltration in LUAD from the TIMER2.0 database. The figure was created using the TIMER2.0 database (http://timer.cistrome.org/).
Figure 9. Correlation of JAML expression with immune infiltration in LUAD from the TIMER2.0 database. The figure was created using the TIMER2.0 database (http://timer.cistrome.org/). 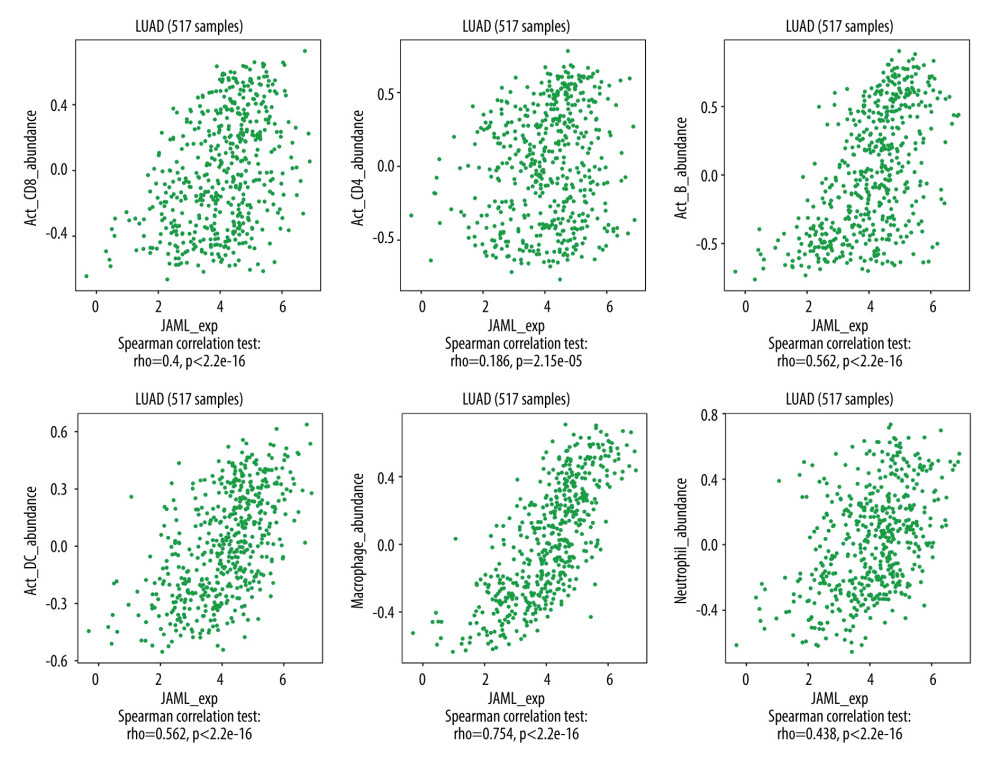 Figure 10. Correlation of JAML expression with immune infiltration in LUAD from the TISIDB. The figure was created using TISIDB (http://cis.hku.hk/TISIDB/index.php).
Figure 10. Correlation of JAML expression with immune infiltration in LUAD from the TISIDB. The figure was created using TISIDB (http://cis.hku.hk/TISIDB/index.php). Tables
Table 1. Baseline characteristics of the LUAD patients.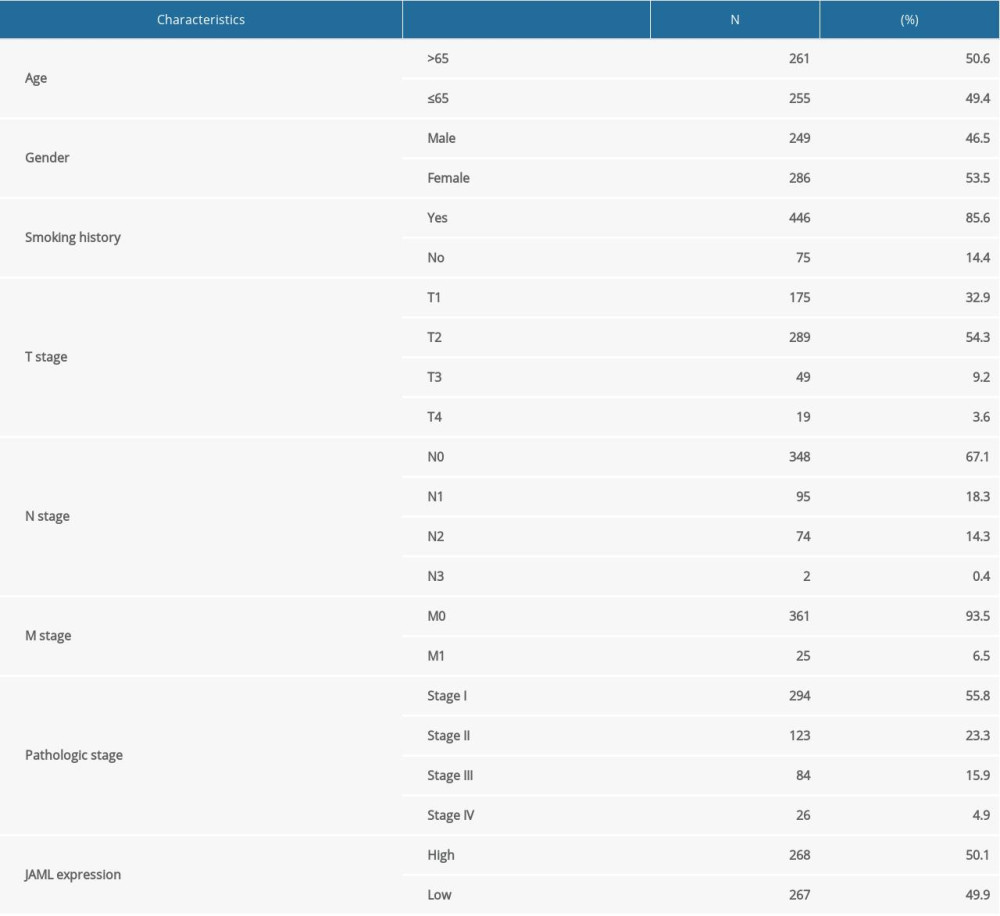 Table 2. Cox regression analysis of JAML and clinicopathologic characteristics with survival outcomes in LUAD.
Table 2. Cox regression analysis of JAML and clinicopathologic characteristics with survival outcomes in LUAD.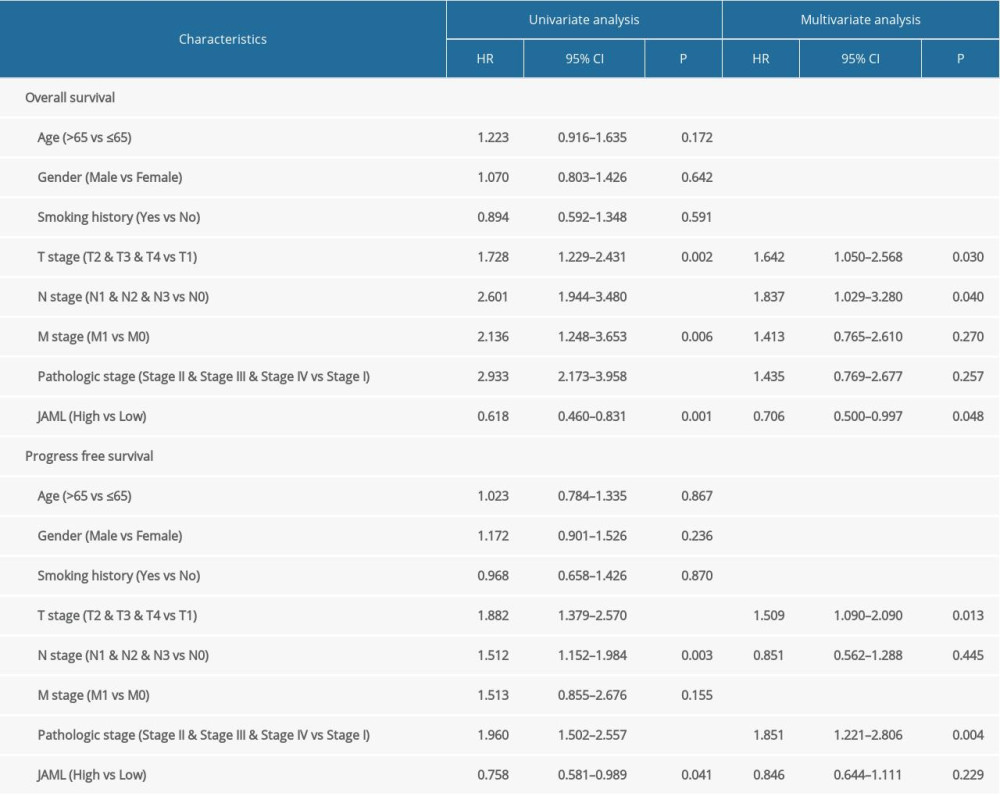 Table 3. Correlation analysis between JAML and markers of immune cells in GEPIA.
Table 3. Correlation analysis between JAML and markers of immune cells in GEPIA.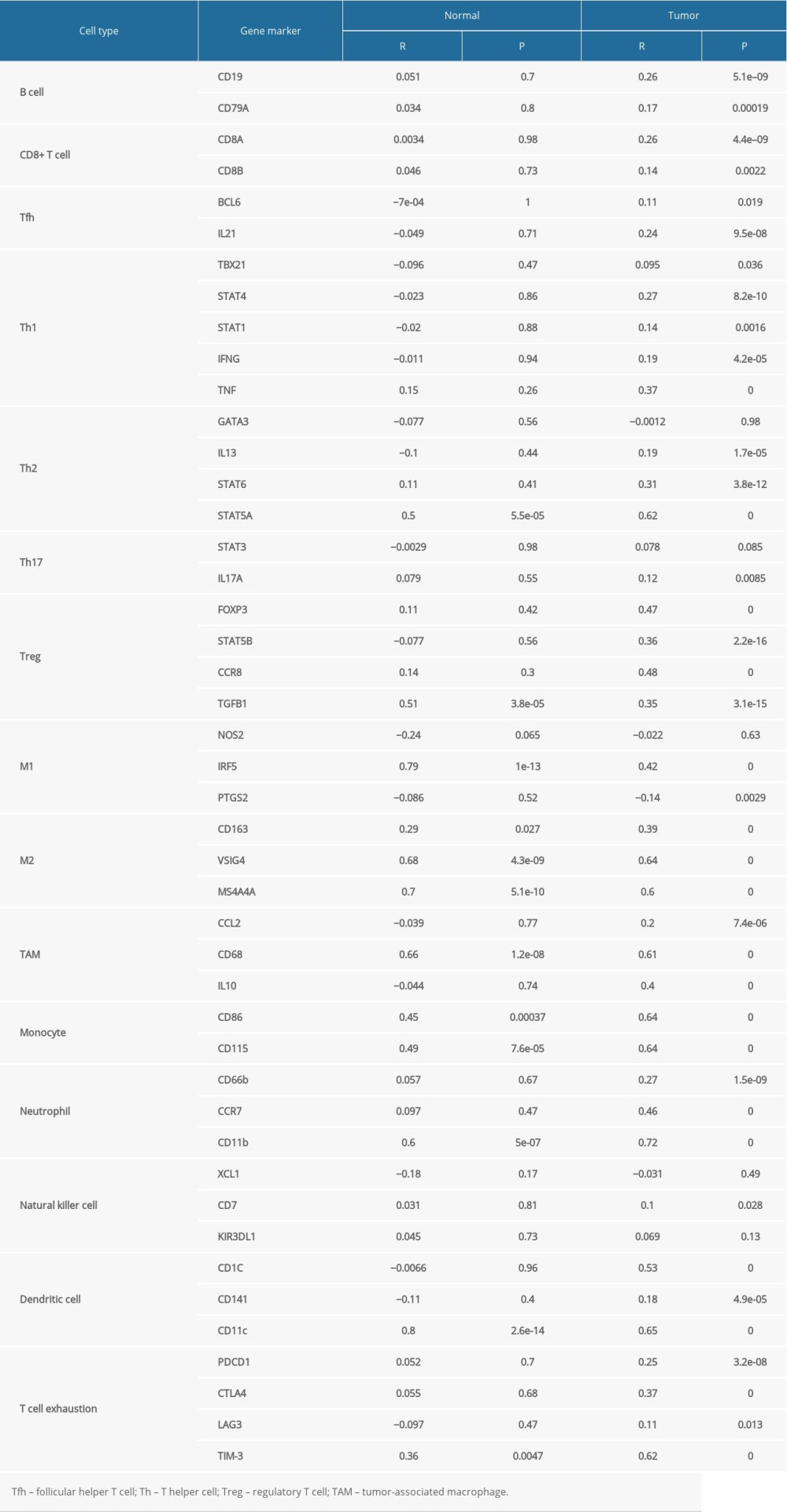 Supplementary Table 1. The top 50 genes that were positively associated with JAML.
Supplementary Table 1. The top 50 genes that were positively associated with JAML.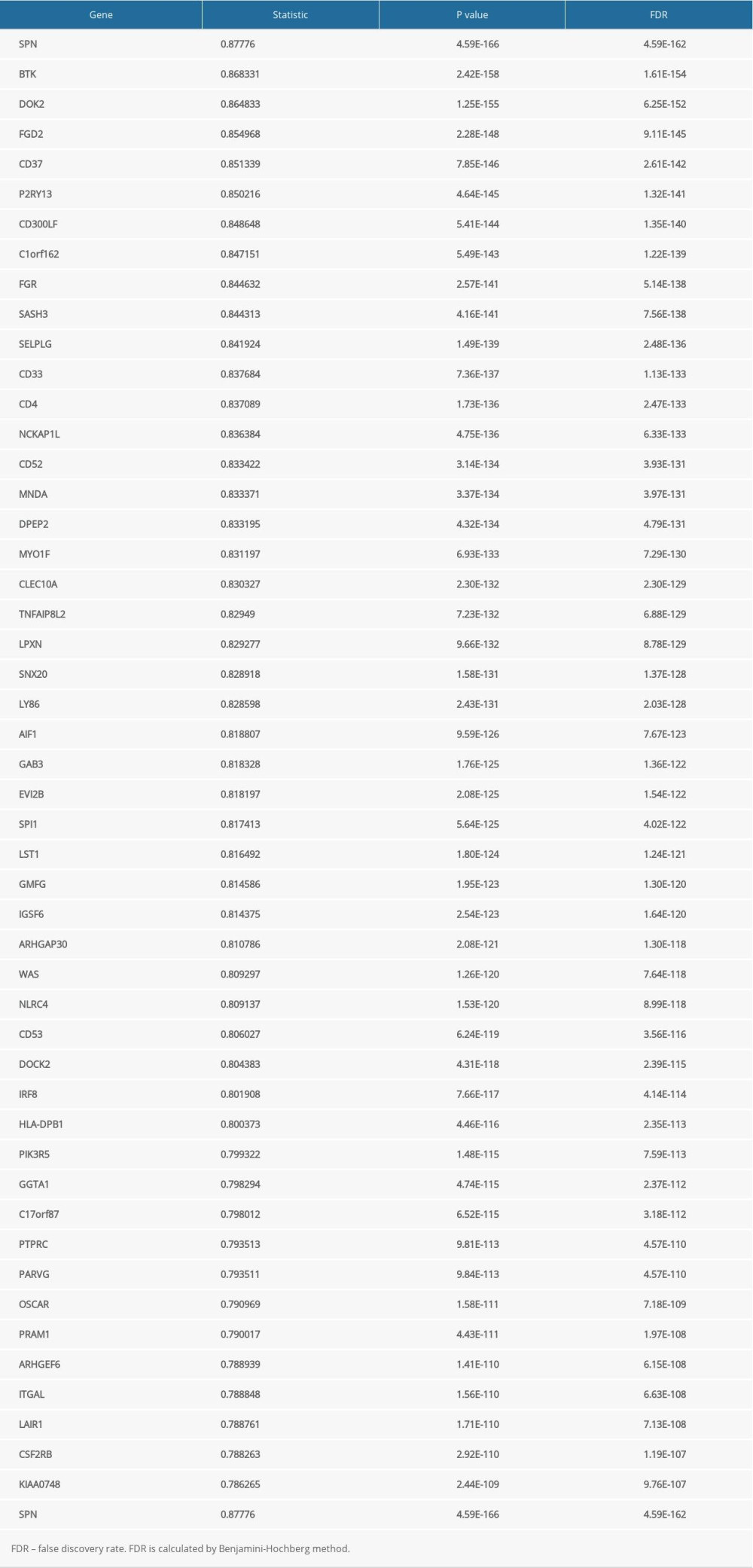 Supplementary Table 2. The top 50 genes that were negatively associated with JAML.
Supplementary Table 2. The top 50 genes that were negatively associated with JAML.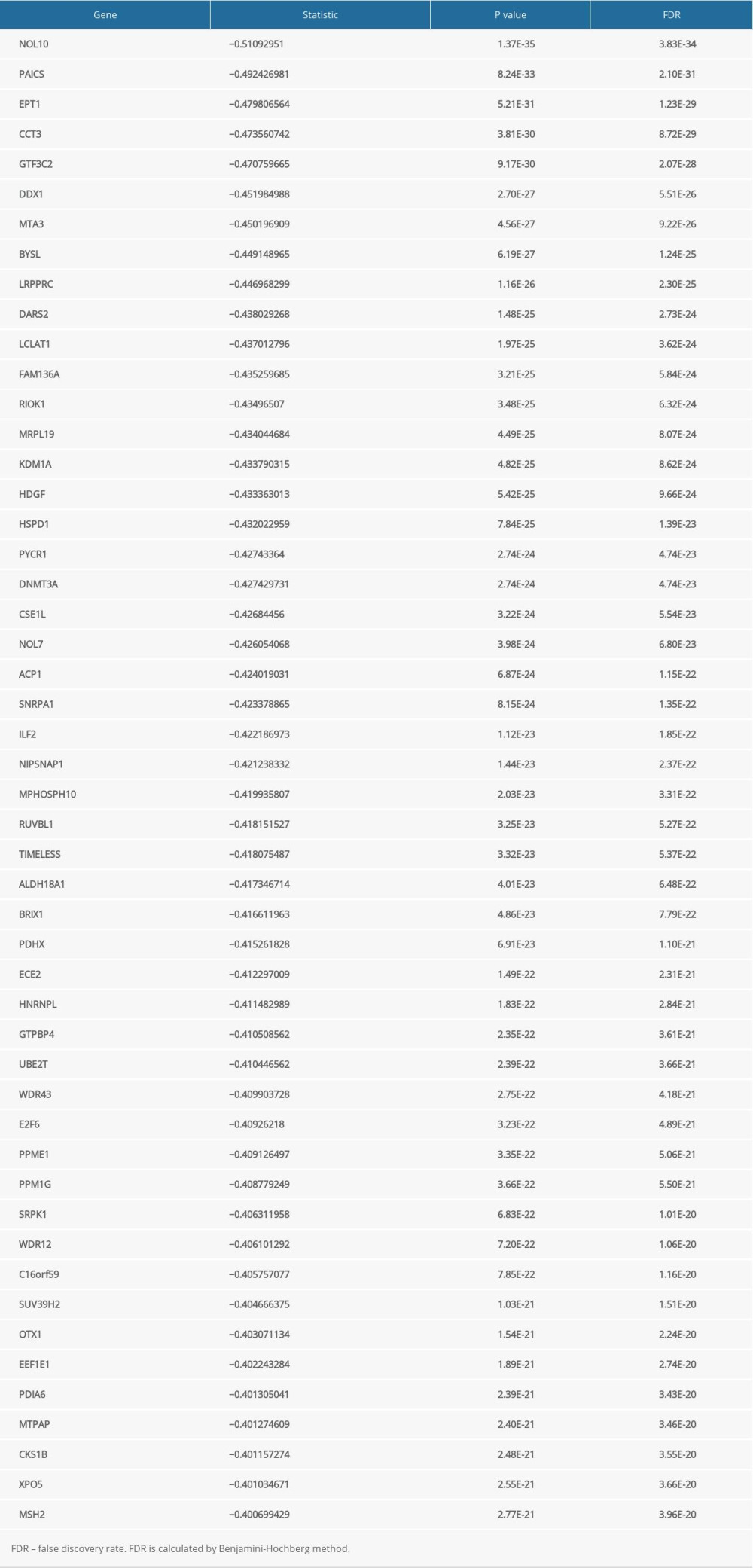
References
1. Siegel RL, Miller KD, Jemal A, Cancer statistics, 2020: Cancer J Clin, 2020; 70(1); 7-30
2. Bray F, Ferlay J, Soerjomataram I, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries: Cancer J Clin, 2018; 68(6); 394-424
3. Travis WD, Brambilla E, Burke AP, Introduction to The 2015 World Health Organization Classification of tumors of the lung, pleura, thymus, and heart: J Thorac Oncol, 2015; 10(9); 1240-42
4. Arbour KC, Riely GJ, Systemic therapy for locally advanced and metastatic non-small cell lung cancer: A review: JAMA, 2019; 322(8); 764-74
5. Piper-Vallillo AJ, Sequist LV, Piotrowska Z, Emerging treatment paradigms for EGFR-mutant lung cancers progressing on osimertinib: A review: J Clin Oncol, 2020 [Online ahead of print]
6. Planchard D, Adjuvant osimertinib in EGFR-mutated non-small-cell lung cancer: N Engl J Med, 2020; 383(18); 1780-82
7. Yang CY, Yang JC, Yang PC, Precision management of advanced non-small cell lung cancer: Annu Rev Med, 2020; 71; 117-36
8. Nadal E, Massuti B, Domine M, Immunotherapy with checkpoint inhibitors in non-small cell lung cancer: Insights from long-term survivors: Cancer Immunol Immunother, 2019; 68(3); 341-52
9. Hegde PS, Chen DS, Top 10 challenges in cancer immunotherapy: Immunity, 2020; 52(1); 17-35
10. Moog-Lutz C, Cavé-Riant F, Guibal FC, JAML, a novel protein with characteristics of a junctional adhesion molecule, is induced during differentiation of myeloid leukemia cells: Blood, 2003; 102(9); 3371-78
11. Weber C, Fraemohs L, Dejana E, The role of junctional adhesion molecules in vascular inflammation: Nat Rev Immunol, 2007; 7(6); 467-77
12. Verdino P, Witherden DA, Havran WL, The molecular interaction of CAR and JAML recruits the central cell signal transducer PI3K: Science (New York, NY), 2010; 329(5996); 1210-14
13. Sanati N, Iancu OD, Wu G, Network-based predictors of progression in head and neck squamous cell carcinoma: Front Genet, 2018; 9; 183
14. Vasaikar SV, Straub P, Wang J, LinkedOmics: Analyzing multi-omics data within and across 32 cancer types: Nucleic Acids Res, 2018; 46(D1); D956-63
15. Franz M, Rodriguez H, Lopes C, GeneMANIA update 2018: Nucleic Acids Res, 2018; 46(W1); W60-64
16. Li T, Fu J, Zeng Z, TIMER2.0 for analysis of tumor-infiltrating immune cells: Nucleic Acids Res, 2020; 48(W1); W509-14
17. Ru B, Wong CN, Tong Y, TISIDB: An integrated repository portal for tumor-immune system interactions: Bioinformatics (Oxford, England), 2019; 35(20); 4200-2
18. Tang Z, Kang B, Li C, GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis: Nucleic Acids Res, 2019; 47(W1); W556-60
19. Hanahan D, Weinberg RA, Hallmarks of cancer: The next generation: Cell, 2011; 144(5); 646-74
20. Schalper KA, Brown J, Carvajal-Hausdorf D, Objective measurement and clinical significance of TILs in non-small cell lung cancer: J Natl Cancer Inst, 2015; 107(3); dju435
21. Paijens ST, Vledder A, de Bruyn M, Tumor-infiltrating lymphocytes in the immunotherapy era: Cell Mol Immunol, 2021; 18(4); 842-59
22. Bazzoni G, The JAM family of junctional adhesion molecules: Curr Opin Cell Biol, 2003; 15(5); 525-30
23. Orlandella FM, Mariniello RM, Iervolino PLC, Junctional adhesion molecule-A is down-regulated in anaplastic thyroid carcinomas and reduces cancer cell aggressiveness by modulating p53 and GSK3 alpha/beta pathways: Mol Carcinog, 2019; 58(7); 1181-93
24. Bonilha CS, Benson RA, Brewer JM, Targeting opposing immunological roles of the junctional adhesion molecule-A in autoimmunity and cancer: Front Immunol, 2020; 11; 602094
25. Liu C, Li X, Shao H, Identification and validation of two lung adenocarcinoma-development characteristic gene sets for diagnosing lung adenocarcinoma and predicting prognosis: Front Genet, 2020; 11; 565206
26. Bosse Y, Li Z, Xia J, Transcriptome-wide association study reveals candidate causal genes for lung cancer: Int J Cancer, 2020; 146(7); 1862-78
27. Verdino P, Witherden DA, Ferguson MS, Molecular insights into γδ T cell costimulation by an anti-JAML antibody: Structure (London, England: 1993), 2011; 19(1); 80-89
28. Witherden DA, Verdino P, Rieder SE, The junctional adhesion molecule JAML is a costimulatory receptor for epithelial gammadelta T cell activation: Science (New York, NY), 2010; 329(5996); 1205-10
29. Roh SE, Jeong Y, Kang MH, Junctional adhesion molecules mediate transendothelial migration of dendritic cell vaccine in cancer immunotherapy: Cancer Lett, 2018; 434; 196-205
30. Kim HR, Park SM, Seo SU, The ratio of peripheral regulatory T cells to Lox-1(+) polymorphonuclear myeloid-derived suppressor cells predicts the early response to anti-PD-1 therapy in patients with non-small cell lung cancer: Am J Respir Crit Care Med, 2019; 199(2); 243-46
Figures
 Figure 1. (A) JAML expression was lower in LUAD tissues than in normal tissues in TCGA database. (B) The ROC curve of JAML expression in LUAD. TPR – true positive rate; FPR – false positive rate. *** P<0.001. The figure was created using R statistical software (version 3.6.3).
Figure 1. (A) JAML expression was lower in LUAD tissues than in normal tissues in TCGA database. (B) The ROC curve of JAML expression in LUAD. TPR – true positive rate; FPR – false positive rate. *** P<0.001. The figure was created using R statistical software (version 3.6.3). Figure 2. Validation of lower JAML expression in LUAD than that in normal tissues in (A) GSE10799, (B) GSE27262, (C) GSE40791, and (D) GSE118370 datasets. ** P<0.01; *** P<0.001. The figure was created using R statistical software (version 3.6.3).
Figure 2. Validation of lower JAML expression in LUAD than that in normal tissues in (A) GSE10799, (B) GSE27262, (C) GSE40791, and (D) GSE118370 datasets. ** P<0.01; *** P<0.001. The figure was created using R statistical software (version 3.6.3). Figure 3. (A) There was no significant difference between JAML expression and age. (B) JAML expression was lower in males than in females. (C) JAML expression was lower in patients with smoking history than in those without smoking history. * P<0.05; ** P<0.01. The figure was created using R statistical software (version 3.6.3).
Figure 3. (A) There was no significant difference between JAML expression and age. (B) JAML expression was lower in males than in females. (C) JAML expression was lower in patients with smoking history than in those without smoking history. * P<0.05; ** P<0.01. The figure was created using R statistical software (version 3.6.3). Figure 4. (A) JAML expression was lower in T2 and T3 stage than in T1 stage, but the difference was not observed in T4 stage. (B) Low JAML expression was associated with positive N stage. (C) There was no significant difference between JAML expression and M stage. (D) Low JAML expression was associated with advanced pathologic stage. ns, P≥0.05; * P<0.05; ** P<0.01; *** P<0.001. The figure was created using R statistical software (version 3.6.3).
Figure 4. (A) JAML expression was lower in T2 and T3 stage than in T1 stage, but the difference was not observed in T4 stage. (B) Low JAML expression was associated with positive N stage. (C) There was no significant difference between JAML expression and M stage. (D) Low JAML expression was associated with advanced pathologic stage. ns, P≥0.05; * P<0.05; ** P<0.01; *** P<0.001. The figure was created using R statistical software (version 3.6.3). Figure 5. Kaplan-Meier survival curves for (A) overall survival (OS) and (B) progression-free survival (PFS) of the LUAD patients with high and low JAML expression level. The figure was created using R statistical software (version 3.6.3).
Figure 5. Kaplan-Meier survival curves for (A) overall survival (OS) and (B) progression-free survival (PFS) of the LUAD patients with high and low JAML expression level. The figure was created using R statistical software (version 3.6.3). Figure 6. Multivariate Cox regression analysis of JAML expression and clinicopathologic characteristics with (A) overall survival (OS) and (B) progression-free survival (PFS) in LUAD. The figure was created using R statistical software (version 3.6.3).
Figure 6. Multivariate Cox regression analysis of JAML expression and clinicopathologic characteristics with (A) overall survival (OS) and (B) progression-free survival (PFS) in LUAD. The figure was created using R statistical software (version 3.6.3). Figure 7. The co-expression genes with JAML in LUAD from the LinkedOmics database. (A, B) Heat maps of top 50 genes positively and negatively correlated with JAML in LUAD, respectively. (C, D) GO annotations and KEGG pathways of JAML in LUAD. The figure was created using the LinkedOmics database (http://www.linkedomics.org).
Figure 7. The co-expression genes with JAML in LUAD from the LinkedOmics database. (A, B) Heat maps of top 50 genes positively and negatively correlated with JAML in LUAD, respectively. (C, D) GO annotations and KEGG pathways of JAML in LUAD. The figure was created using the LinkedOmics database (http://www.linkedomics.org). Figure 8. Protein–protein interaction network of JAML. The figure was created using the GeneMANIA database (http://genemania.org).
Figure 8. Protein–protein interaction network of JAML. The figure was created using the GeneMANIA database (http://genemania.org). Figure 9. Correlation of JAML expression with immune infiltration in LUAD from the TIMER2.0 database. The figure was created using the TIMER2.0 database (http://timer.cistrome.org/).
Figure 9. Correlation of JAML expression with immune infiltration in LUAD from the TIMER2.0 database. The figure was created using the TIMER2.0 database (http://timer.cistrome.org/). Figure 10. Correlation of JAML expression with immune infiltration in LUAD from the TISIDB. The figure was created using TISIDB (http://cis.hku.hk/TISIDB/index.php).
Figure 10. Correlation of JAML expression with immune infiltration in LUAD from the TISIDB. The figure was created using TISIDB (http://cis.hku.hk/TISIDB/index.php). Tables
 Table 1. Baseline characteristics of the LUAD patients.
Table 1. Baseline characteristics of the LUAD patients. Table 2. Cox regression analysis of JAML and clinicopathologic characteristics with survival outcomes in LUAD.
Table 2. Cox regression analysis of JAML and clinicopathologic characteristics with survival outcomes in LUAD. Table 3. Correlation analysis between JAML and markers of immune cells in GEPIA.
Table 3. Correlation analysis between JAML and markers of immune cells in GEPIA. Table 1. Baseline characteristics of the LUAD patients.
Table 1. Baseline characteristics of the LUAD patients. Table 2. Cox regression analysis of JAML and clinicopathologic characteristics with survival outcomes in LUAD.
Table 2. Cox regression analysis of JAML and clinicopathologic characteristics with survival outcomes in LUAD. Table 3. Correlation analysis between JAML and markers of immune cells in GEPIA.
Table 3. Correlation analysis between JAML and markers of immune cells in GEPIA. Supplementary Table 1. The top 50 genes that were positively associated with JAML.
Supplementary Table 1. The top 50 genes that were positively associated with JAML. Supplementary Table 2. The top 50 genes that were negatively associated with JAML.
Supplementary Table 2. The top 50 genes that were negatively associated with JAML. In Press
15 Apr 2024 : Laboratory Research
The Role of Copper-Induced M2 Macrophage Polarization in Protecting Cartilage Matrix in OsteoarthritisMed Sci Monit In Press; DOI: 10.12659/MSM.943738
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








