31 May 2021: Clinical Research
Association of High-Mobility Group Box 1 (HMGB1) Gene Polymorphisms with Susceptibility and Better Survival Prognosis in Chinese Han Neonatal Necrotizing Enterocolitis
Huiling Cao1ABCEF, Defeng Guo2CEF*DOI: 10.12659/MSM.930015
Med Sci Monit 2021; 27:e930015
Abstract
BACKGROUND: High-mobility group box 1 (HMGB1) plays a crucial role in a variety of diseases, including neonatal necrotizing enterocolitis (NEC). The purpose of this study was to investigate the association of HMGB1 gene single-nucleotide polymorphisms (SNPs) with susceptibility and survival prognosis in Chinese Han neonates with NEC.
MATERIAL AND METHODS: The HMGB1 gene rs1360485, rs1045411, and rs2249825 site SNPs were genotyped in all participants. The mRNA expression of serum HMGB1 was examined using quantitative reverse transcription-polymerase chain reaction. The correlation of the HMGB1 rs1360485 SNP with NEC neonatal survival prognosis was evaluated by univariate analysis and logistic multivariate regression analysis.
RESULTS: The TC and CC genotype and C allele distribution frequencies of the rs1360485 SNP were lower in the NEC group, and the differences were statistically significant (all P<0.05). Individuals carrying the TC and CC genotype or C allele had a low risk of being affected by NEC. However, the genotype and allele distributions of rs1045411 and rs2249825 were not significantly different between the patient and control groups (P>0.05). NEC neonates with HMGB1 gene rs1360485 site mutations had lower mRNA levels of serum HMGB1 than those with rs1360485 site wild-type, and the rs1360485 genotypes TC and CC could independently predict better survival outcomes in NEC neonates.
CONCLUSIONS: This study demonstrated that the rs1360485 SNP of the HMGB1 gene is associated with susceptibility of NEC in neonates, and the rs1360485 genotypes TC and CC may affect HMGB1 expression and are associated with the survival prognosis of neonates with NEC.
Keywords: Disease Susceptibility, Enterocolitis, HMGB Proteins, Polymorphism, Single Nucleotide, Disease Progression, Enterocolitis, Necrotizing, Genetic Predisposition to Disease, HMGB1 Protein, Infant, Newborn, Risk Factors
Background
Necrotizing enterocolitis (NEC) is a severe intestinal inflammatory disease that mainly affects preterm infants and remains a major cause of neonatal morbidity and mortality [1,2]. NEC is often associated with premature birth, ischemia, formula feeding, antibiotics, and some risk factors that predict intestinal mucosal damage or intestinal abnormalities [3]. However, the presence of these risk factors does not necessarily lead to an increased incidence or severity in neonates with NEC [3], since NEC occurs only in certain newborns even with exposure to similar environmental conditions or interference. Although the etiology of NEC is complex, and the exact pathogenesis of the disease remains unclear, an underlying genetic predisposition to NEC has been increasingly recognized in recent years [1,4]. Therefore, studying candidate genes associated with the development and progression of NEC may help explain the molecular mechanism of NEC.
High-mobility group box 1 (HMGB1) protein is a nuclear protein ubiquitously present in almost all cell types [5]. In addition to having intracellular functions, HMGB1 can be released extracellularly as an advanced inflammatory mediator, which can be actively secreted by activated macrophages, monocytes, and natural killer cells and passively released by necrotic cells [6,7]. It has been reported that HMGB1 could stimulate the production of pro-inflammatory cytokines, and that these cytokines are hypothetical mediators of NEC-associated intestinal inflammation [8]. Several studies have revealed that HMGB1 is significantly increased in neonates with NEC [9–11], participates in the development of NEC, and plays a crucial role in the pathological mechanism of NEC. For example, a study demonstrated that HMGB1 is upregulated in the progression of NEC, and that the inhibition of HMGB1 attenuates intestinal inflammation in NEC [12]. The increased expression of HMGB1 in the intestine plays an important role in human NEC [13]. Zamora et al found that the inhibition of HMGB1 expression can limit intestinal damage in experimental NEC [8].
There are multiple variants in
Consequently, this study explored the association of the
Material and Methods
PATIENTS AND SAMPLE COLLECTION:
A total of 258 neonates who were admitted to Weifang People’s Hospital from 2011 to 2018 with NEC were recruited as a case group. The inclusion criteria were (1) Han ethnicity, (2) gestational age <37 weeks, and (3) NEC was diagnosed according to the criteria originally defined by Bell et al, and subsequently modified by Walsh and Kliegman [19]. Exclusion criteria were (1) congenital intestinal malformations, (2) congenital genetic metabolic diseases, and (3) combined NEC-unrelated gastrointestinal inflammation or infectious diseases. The neonates with NEC included 154 boys and 104 girls, with a gestational age of 34.1±2.2 weeks and a birth weight of 2267±381 g. According to NEC staging criteria [20], they were divided into stage II (161 cases) and stage III (97 cases). In addition, 180 newborns matched with the NEC case group in terms of gestational age, birth weight, and sex were enrolled as a control group during the same period, and the inclusion criteria were (1) Han ethnicity, (2) gestational age <37 weeks, (3) no NEC, and (4) no severe underlying diseases. The control group consisted of 98 boys and 82 girls, with a gestational age of 34.6±2.3 weeks and a birth weight of 2332±408 g. Our study was approved by the Ethics Committee of Weifang People’s Hospital (approval number: 110239), and we obtained informed consent from the guardians of the subjects for the use of serum samples and subsequent analyses. No consanguinity was found among the participants.
An amount of 2 mL of peripheral venous blood was collected from each participant and anticoagulated with 5% ethylene diamine tetra acetic acid. Genomic DNA (gDNA) was extracted with the Takara Genomic DNA extraction kit (Beijing Boiteke Corporation, Beijing, China) and then stored at −20°C for genotyping. In addition, 2-mL samples of blood from the NEC neonates and control newborns were collected without anticoagulant; the blood was separated by centrifugation to obtain the serum and stored at −80°C for further use.
DETERMINATION OF SNPS:
The sequences of HMGB1 SNP sites rs1045411, rs2249825, and rs1360485 were amplified by polymerase chain reaction (PCR) using gDNA as a template. The primer fragments used for amplification were designed by Primer Premier 5.0 and synthesized by Shanghai Sangon Biotech Co., Ltd. (Table 1). The PCR process was as follows: 95°C for 10 min; 35 cycles of 95°C for 30 s, 60°C for 3 s, and 72°C for 4 s; final extension for 10 min at 72°C; and storing at 4°C. The resulting PCR products were purified and sent to Shanghai Biotech Bioengineering Technology Co., Ltd. for sequencing.
RNA EXTRACTION AND QUANTITATIVE REVERSE TRANSCRIPTION PCR:
RNA was extracted from serum using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). A PrimeScript RT reagent kit (TaKaRa, Japan) was used to synthesize cDNA from the obtained RNA.
Serum
STATISTICAL ANALYSIS:
Hardy-Weinberg equilibrium (HWE) was analyzed for each SNP in the case and control groups to evaluate the quality of the study samples. The distribution differences of genotypes and alleles between the 2 groups were compared by the chi-squared test. The correlation of
Results
HWE TEST RESULTS:
As shown in Table 2, the genotype distributions of the HMGB1 gene rs1045411, rs2249825, and rs1360485 SNPs were in accordance with the HWE in the case and control groups (all P>0.05), revealing the representativeness of our study population.
:
The present study used the OR and 95% CI to reflect the association of HMGB1 gene SNPs with neonatal NEC susceptibility to evaluate the influence of genotype and allele frequencies on the onset of NEC. As shown in Table 2, no significant difference was observed for the rs1045411 SNP between the case and control groups. The GG, GA, and AA genotype frequencies were 66.7%, 28.3%, 5.0% in the NEC case group, and 67.8%, 28.3%, 3.9% in the control group, respectively. The G and A allele frequencies were 80.8% and 19.2% in the NEC case group and 81.9% and 18.1% in the control group, respectively, but none of the distribution differences were significant (all P>0.05). Additionally, no significant difference in the rs2249825 SNP of the HMGB1 gene was found between the NEC case and control groups. The CC, CG, and GG genotype frequencies were 72.1%, 24.8%, and 3.1% in the NEC case group and 73.9%, 22.8%, and 3.3% in the control group, respectively. The C and G allele frequencies were 84.5% and 15.5% in the NEC case group and 85.3% and 14.7% in the control group, respectively, but none of the distribution differences were significant (all P>0.05). The results demonstrated that there might be a lack of association between the HMGB1 gene rs1045411 and rs2249825 SNPs and neonatal NEC susceptibility in the Chinese Han neonates.
For the rs1360485 SNP, the frequency of the TC genotype in the NEC case group was significantly lower than that of the control group (34.1% vs 41.7%), and individuals carrying the TC genotype had a lower risk of developing NEC than did TT genotype carriers (OR 0.661, 95% CI 0.443–0.947,
:
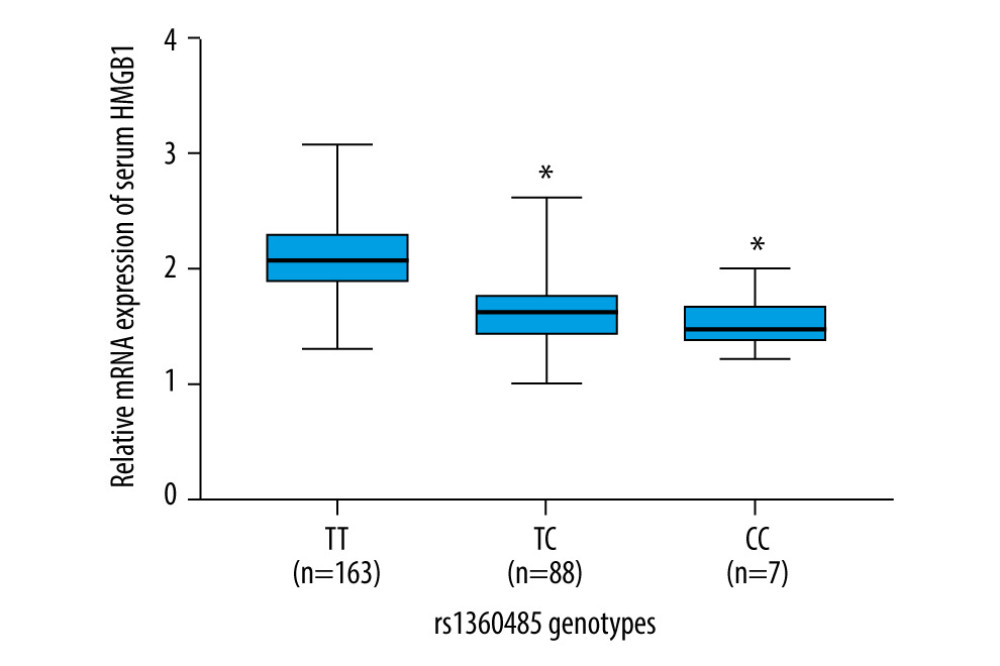
We used the qRT-PCR method to detect serum HMGB1 mRNA expression in NEC neonates. As shown in Figure 1, NEC patients with rs1360485 locus mutations (TC or CC) had significantly lower mRNA expression of serum HMGB1 compared with patients with wild-type (TT) (all P<0.05).
:
We analyzed the factors related with the survival prognosis of NEC (Table 3). NEC neonates were divided into survival (n=197) and death (n=61) groups. The results indicated that metabolic acidosis, neonatal scleroderma, intestinal perforation, diffuse peritonitis, thrombocytopenia, leukocyte disorder, and rs1360485 genotypes TC and CC were associated with survival prognosis of NEC neonates (all P<0.01). However, there was no association between respiratory distress syndrome and survival prognosis of NEC neonates (P>0.05). Further, multivariate logistic regression analysis (Table 4) showed that metabolic acidosis, intestinal perforation, diffuse peritonitis, thrombocytopenia, and rs1360485 genotypes TC and CC (all P<0.05) were independently predictive of survival outcomes in patients with NEC. These results suggested that rs1360485 genotypes TC and CC can independently predict better survival outcomes in patients with NEC.
Discussion
NEC is a serious gastrointestinal disease that occurs in the neonatal period and has a high mortality rate [21]. Neonates with NEC initially present with abdominal distension, intolerance to feeding, and bloody stools and often develop systemic disease involving multiple organs and shock [3,22]. Although epidemiological and other basic studies have greatly improved the understanding of NEC, its exact etiology is still unclear, and there is currently no fully effective prevention or treatment [23]. Thus, exploring the pathogenesis of NEC has become a hot spot of clinical research. HMGB1 can acts as a pleiotropic cytokine and is involved in the pathology of many diverse immune-mediated diseases [24]. When HMGB1 protein is exocrine, it can accelerate the development of NEC. Many data have indicated that inhibiting the expression of the
In the present study, we used the rs1360485, rs2249825, and rs1045411 SNPs in the
However, the results of our study did not find correlations between the rs1045411 and rs2249825 SNPs of the
There is increasing evidence that alterations in
There are some limitations in this study. Our study sample was selected from the same hospital, which may have caused bias in our selection of data and reduced the statistical validity of comparing differences between the 2 groups. Also, the sample size of our study was not large enough, which may have affected the accuracy of the statistical analysis.
Conclusions
In summary, our findings indicated that the rs1360485 locus SNP of the
Tables
Table 1. Primer sequences of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms.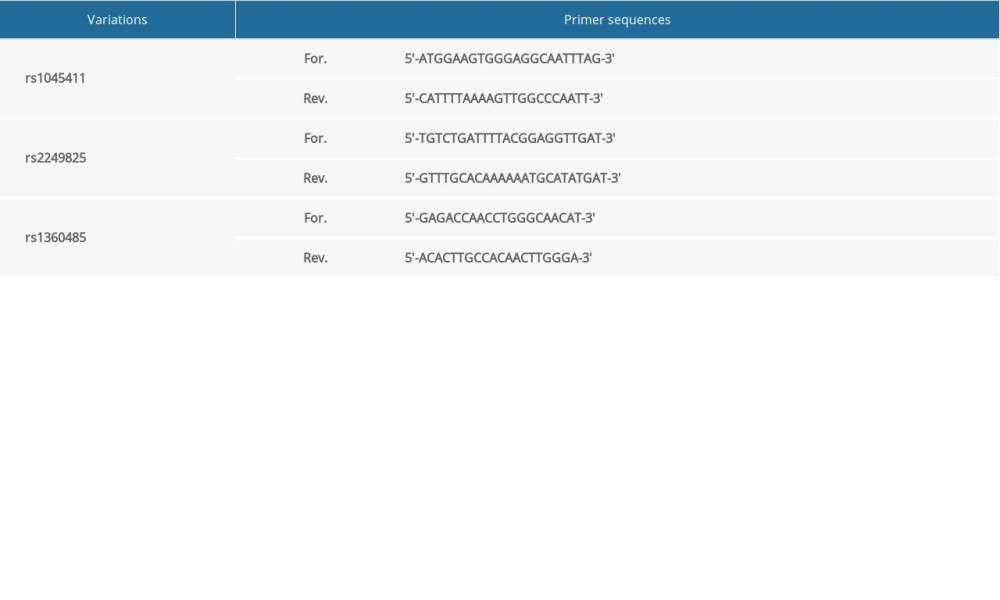 Table 2. Genotype and allele distributions of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms in case and control groups.
Table 2. Genotype and allele distributions of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms in case and control groups.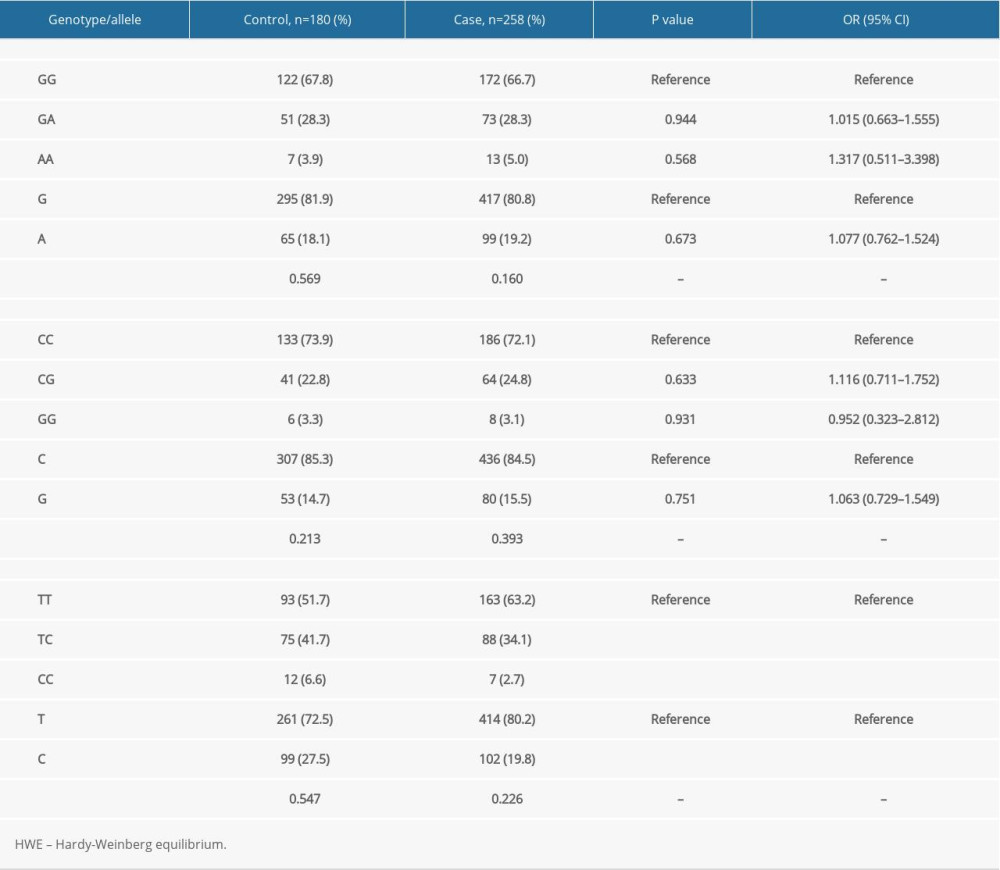 Table 3. Univariate analysis for factors that were related with the survival prognosis of necrotizing enterocolitis (NEC).
Table 3. Univariate analysis for factors that were related with the survival prognosis of necrotizing enterocolitis (NEC).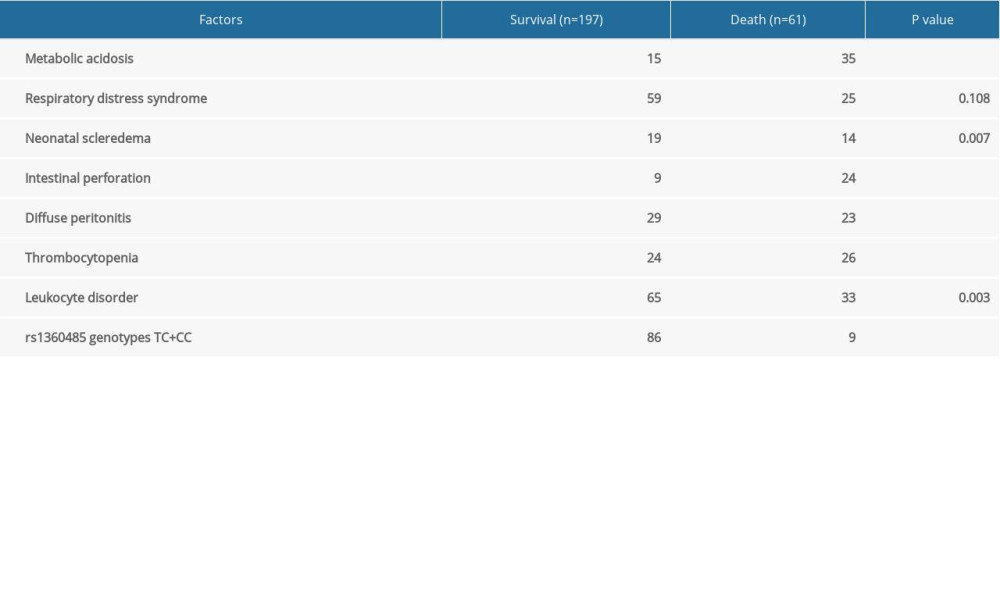 Table 4. Multivariate logistic regression analysis for the factors that independently predict survival outcomes in necrotizing enterocolitis (NEC) patients.
Table 4. Multivariate logistic regression analysis for the factors that independently predict survival outcomes in necrotizing enterocolitis (NEC) patients.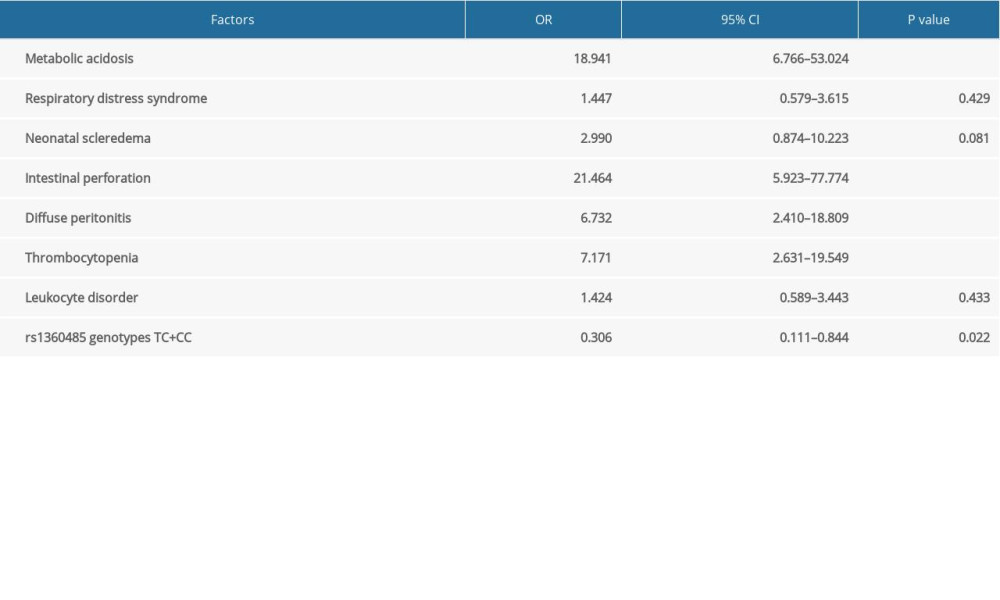
References
1. Cuna A, George L, Sampath V, Genetic predisposition to necrotizing enterocolitis in premature infants: Current knowledge, challenges, and future directions: Semin Fetal Neonat Med, 2018; 23; 387-93
2. Bellodas Sanchez J, Kadrofske M, Necrotizing enterocolitis: Neurogastroenterol Motil, 2019; 31; e13569
3. Sampath V, Bhandari V, Berger J, A functional ATG16L1 (T300A) variant is associated with necrotizing enterocolitis in premature infants: Pediatr Res, 2017; 81; 582-88
4. Cuna A, Sampath V, Genetic alterations in necrotizing enterocolitis: Semin Perinatol, 2017; 41; 61-69
5. Yang H, Antoine DJ, Andersson U, Tracey KJ, The many faces of HMGB1: Molecular structure-functional activity in inflammation, apoptosis, and chemotaxis: J Leukoc Biol, 2013; 93; 865-73
6. Chen Q, Guan X, Zuo X, The role of high mobility group box 1 (HMGB1) in the pathogenesis of kidney diseases: Acta Pharm Sin B, 2016; 6; 183-88
7. Venereau E, De Leo F, Mezzapelle R, HMGB1 as biomarker and drug target: Pharmacol Res, 2016; 111; 534-44
8. Zamora R, Grishin A, Wong C, High-mobility group box 1 protein is an inflammatory mediator in necrotizing enterocolitis: Protective effect of the macrophage deactivator semapimod: Am J Physiol Gastrointest Liver Physiol, 2005; 289; G643-52
9. Li B, Saka R, Takama Y, Recombinant human soluble thrombomodulin reduces the severity and incidence of necrotizing enterocolitis in a newborn rat model: Surg Today, 2019; 49; 971-76
10. Downard CD, Grant SN, Matheson PJ, Altered intestinal microcirculation is the critical event in the development of necrotizing enterocolitis: J Pediatr Surg, 2011; 46; 1023-28
11. Nino DF, Zhou Q, Yamaguchi Y, Cognitive impairments induced by necrotizing enterocolitis can be prevented by inhibiting microglial activation in mouse brain: Sci Transl Med, 2018; 10; eaan0237
12. Yu R, Jiang S, Tao Y, Inhibition of HMGB1 improves necrotizing enterocolitis by inhibiting NLRP3 via TLR4 and NF-kappaB signaling pathways: J Cell Physiol, 2019; 234; 13431-38
13. Dai S, Sodhi C, Cetin S, Extracellular high mobility group box-1 (HMGB1) inhibits enterocyte migration via activation of Toll-like receptor-4 and increased cell-matrix adhesiveness: J Biol Chem, 2010; 285; 4995-5002
14. Qiu P, Wang L, Ni J, Zhang Y, Associations between HMGB1 gene polymorphisms and susceptibility and clinical outcomes in Chinese Han sepsis patients: Gene, 2019; 687; 23-29
15. Song W, Tan H, Wang S, Association of high mobility group box protein B1 gene polymorphisms with pneumonia susceptibility and severity: Genet Test Mol Biomarkers, 2019; 23; 3-11
16. Lin CW, Chou YE, Yeh CM, A functional variant at the miRNA binding site in HMGB1 gene is associated with risk of oral squamous cell carcinoma: Oncotarget, 2017; 8; 34630-42
17. Wu HH, Liu YF, Yang SF, Association of single-nucleotide polymorphisms of high-mobility group box 1 with susceptibility and clinicopathological characteristics of uterine cervical neoplasia in Taiwanese women: Tumour Biol, 2016 [Online ahead of print]
18. Jin H, Wu J, Yang Q, High mobility group box 1 protein polymorphism affects susceptibility to recurrent pregnancy loss by up-regulating gene expression in chorionic villi: J Assist Reprod Genet, 2015; 32; 1123-28
19. Sharma R, Hudak ML, A clinical perspective of necrotizing enterocolitis: past, present, and future: Clin Perinatol, 2013; 40; 27-51
20. Walsh MC, Kliegman RM, Necrotizing enterocolitis: treatment based on staging criteria: Pediatr Clin North Am, 1986; 33; 179-201
21. Zhou W, Yuan W, Huang L, Association of neonatal necrotizing enterocolitis with myeloid differentiation-2 and GM2 activator protein genetic polymorphisms: Mol Med Rep, 2015; 12; 974-80
22. Gordon P, Christensen R, Weitkamp JH, Maheshwari A, Mapping the new world of necrotizing enterocolitis (NEC): review and opinion: EJ Neonatol Res, 2012; 2; 145-72
23. Jain L, Necrotizing enterocolitis prevention: Art or science?: Clin Perinatol, 2013; 40; xiii-xv
24. Deng CQ, Deng GH, Wang YM, HMGB1 gene polymorphisms in patients with chronic hepatitis B virus infection: World J Gastroenterol, 2013; 19; 5144-49
25. Tang Y, Duan J, Wang Y, Yuan L, Associations of HMGB1 gene polymorphisms with risk of coal workers’ pneumoconiosis susceptibility in Chinese Han population: Inhal Toxicol, 2020; 32; 170-76
26. Chou YE, Yang PJ, Lin CY, The impact of HMGB1 polymorphisms on prostate cancer progression and clinicopathological characteristics: Int J Environ Res, 2020; 17; 7247
27. Hung SC, Wang SS, Li JR, Effect of HMGB1 polymorphisms on urothelial cell carcinoma susceptibility and clinicopathological characteristics: Int J Med Sci, 2018; 15; 1731-36
28. Wang JX, Yu HL, Bei SS, Association of HMGB1 gene polymorphisms with risk of colorectal cancer in a chinese population: Med Sci Mon, 2016; 22; 3419-25
29. Wu YL, Chien MH, Chou YE, Association of EGFR mutations and HMGB1 genetic polymorphisms in lung adenocarcinoma patients: J Cancer, 2019; 10; 2907-14
30. Wang LH, Wu MH, Chen PC, Prognostic significance of high-mobility group box protein 1 genetic polymorphisms in rheumatoid arthritis disease outcome: Int J Med Sci, 2017; 14; 1382-88
31. Huang BF, Tzeng HE, Chen PC, HMGB1 genetic polymorphisms are biomarkers for the development and progression of breast cancer: Int J Med Sci, 2018; 15; 580-86
32. Bao G, Qu F, He L, Prognostic significance of tag SNP rs1045411 in HMGB1 of the aggressive gastric cancer in a Chinese population: PLoS One, 2016; 11; e0154378
Tables
 Table 1. Primer sequences of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms.
Table 1. Primer sequences of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms. Table 2. Genotype and allele distributions of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms in case and control groups.
Table 2. Genotype and allele distributions of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms in case and control groups. Table 3. Univariate analysis for factors that were related with the survival prognosis of necrotizing enterocolitis (NEC).
Table 3. Univariate analysis for factors that were related with the survival prognosis of necrotizing enterocolitis (NEC). Table 4. Multivariate logistic regression analysis for the factors that independently predict survival outcomes in necrotizing enterocolitis (NEC) patients.
Table 4. Multivariate logistic regression analysis for the factors that independently predict survival outcomes in necrotizing enterocolitis (NEC) patients. Table 1. Primer sequences of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms.
Table 1. Primer sequences of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms. Table 2. Genotype and allele distributions of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms in case and control groups.
Table 2. Genotype and allele distributions of the HMGB1 gene rs1045411, rs2249825, and rs1360485 polymorphisms in case and control groups. Table 3. Univariate analysis for factors that were related with the survival prognosis of necrotizing enterocolitis (NEC).
Table 3. Univariate analysis for factors that were related with the survival prognosis of necrotizing enterocolitis (NEC). Table 4. Multivariate logistic regression analysis for the factors that independently predict survival outcomes in necrotizing enterocolitis (NEC) patients.
Table 4. Multivariate logistic regression analysis for the factors that independently predict survival outcomes in necrotizing enterocolitis (NEC) patients. In Press
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
06 Mar 2024 : Clinical Research
Prevalence and Variation of Medical Comorbidities in Oral Surgery Patients: A Retrospective Study at Jazan ...Med Sci Monit In Press; DOI: 10.12659/MSM.943884
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952