05 August 2021: Database Analysis
N6-Methyladenosine Regulators Are Involved in the Progression of and Have Clinical Impact on Breast Cancer
Yanni Song12BCEFG, Chaojing Zheng3C, Yangbao Tao3D, Rui Huang3E, Qian Zhang4AE*DOI: 10.12659/MSM.929615
Med Sci Monit 2021; 27:e929615
Abstract
BACKGROUND: N6-methyladenosine (m⁶A) modification has been widely studied in various cancers, and m6A regulators, such as METTL3, METTL14, WTAP, and YTHDF1, play crucial roles in breast cancer. However, a comprehensive study of m6A regulators in breast cancer is still lacking.
MATERIAL AND METHODS: Expression data of m⁶A regulators and clinicopathological information were acquired from The Cancer Genome Atlas (TCGA) program. Protein interaction was collected from the STRING database. Data on tumor purity and correlation among m6A regulators were obtained from the TIMER database. LASSO, consensus clustering, and gene set enrichment analysis (GSEA) were used to evaluate the role of m⁶A regulators. Moreover, the prognostic value of m⁶A-related genomic targets in breast cancer was analyzed by Kaplan-Meier analysis and Cox regression models.
RESULTS: We found most m⁶A regulators were associated with key clinicopathological parameters, such as tumor staging, Nottingham prognostic index (NPI), and cellularity. Also, consensus clustering analysis-based grouping could effectively predict patients’ overall survival. Correlation analysis also showed that these regulators interacted with each other. Patients were further split into a high-risk group and low-risk group based on Cox and LASSO analysis. High-risk patients had a significantly worse overall survival than did low-risk patients. Moreover, AKT1 and MYC were enriched in patients in the high-risk group, according to GSEA analysis. The patients in the high-risk group also displayed resistance to chemoradiotherapy or hormone therapy.
CONCLUSIONS: The m⁶A regulators are critical participants in the development and progression of breast cancer and are likely to be used to predict prognosis and develop treatment strategies.
Keywords: Breast Neoplasms, chemoradiotherapy, Methylation, Cell Cycle Proteins, Gene Regulatory Networks, Methyltransferases, Predictive Value of Tests, Protein Interaction Maps, RNA Splicing Factors, RNA-Binding Proteins, Risk Assessment
Background
Breast cancer is one of the most common and lethal malignancies in women worldwide. It is estimated that about 1 806 590 new cases and 606 520 deaths will occur in 2020 [1], and therefore immediate action is needed. Although enormous progress has been achieved in the past decades, with the development of surgical skills, diagnosis, and treatment modalities, many questions remain to be answered, such as specific risk factors of breast cancer, limits of immunotherapy, and identification of patients in need of treatment [2].
Epigenetics, mainly including histone modification, chromosome remodeling, acetylation, and methylation, could influence genes’ transcription and translation without changing sequences of DNA [3]. Epigenetics is also involved in the carcinogenesis, development, and prognosis of breast cancer [4]. As a notable methylation form, N6-methyladenosine (m6A) was first discovered and studied in the 1970s and since then has been viewed as the main methylation modification in eukaryotic cells [5]. The m6A regulators are a group of molecules that regulate m6A by adding, erasing, or recognizing the methyl group at the N6 position of adenosine. The m6A process depends on these regulators: methyltransferases (“writers”), demethylases (“erasers”), and binding proteins (“readers”). “Writers” install m6A units to RNA and are mainly composed of Wilms tumor 1- associated protein (WTAP) [6], methyltransferase-like 3 (METTL3) [7], and METTL14 [7]. “Erasers” can remove the m6A unit from molecules, and consist of α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5) [8] and fat-mass and obesity-associated protein (FTO) [9]. The modification of m6A is recognized by the “readers”, which contain heterogeneous nuclear ribonucleoproteins (HNRNPs), YT521-B homology (YTH) family members, and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) [10]. In addition to the above, METTL16, RBM15, KIAA1429, RBM15B, EIF3, ZC3H13, and CBLL1 were included in our analysis. In all, 20 m6A regulators were selected in our study, based on previous research and reviews [11,12].
A growing number of studies are focusing on the role of m6A in breast cancer. Data have shown that m6A modification regulates breast cancer progression [13] and is correlated with prognosis [14]. The knockdown of METTL3 could decrease the m6A methylation level, and therefore inhibit breast cancer proliferation and tumor growth [13]. It has been demonstrated that upregulated YTHDF1, YTHDF3, and KIAA1429 can predict poor overall survival (OS). Overexpression of YTHDF3 is an independent factor for OS in patients with breast cancer [14]. To fully understand the role of m6A in breast cancer, we used data from The Cancer Genome Atlas (TCGA) to comprehensively analyze RNA expression patterns and their potential relevance to clinical characteristics in breast cancer. Our goal was to provide new evidence about the potential mechanisms of m6A regulators and their involvement in the development and progression of breast cancer and to clarify how this evidence could be used as a tool to predict prognosis of breast cancer patients.
Material and Methods
DATA COLLECTION AND ANALYSIS:
The R package getFirehoseData was used to acquire breast cancer data from the TCGA dataset (Breast Cancer, METABRIC, Nature 2012 & Nature Commun 2016). A total of 1904 cases with gene expression and clinical data were included in our analysis. Clinical information and RNA expression of breast cancer patients were downloaded and processed with R version 3.6.1.
Protein interaction data were downloaded from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database [15] (https://string-db.org/) and then analyzed with Cytoscape (https://cytoscape.org/).
The Tumor Immune Estimation Resource (TIMER) [16] (https://cistrome.shinyapps.io/timer/) was adopted to obtainthe correlation among different m6A regulators after adjusting by tumor purity.
GENE SET ENRICHMENT ANALYSIS:
Gene set enrichment analysis (GSEA) (http://software.broadinstitute.org/gsea/index.jsp) was performed to determine whether a series of a previously defined set of genes were enriched in different biological groups [17]. Termswith P<0.05 and FDR<0.25 were identified. The hallmark gene set “h.all.v6.0.symbols.gmt” was used in this study.
LEAST ABSOLUTE SHRINKAGE AND SELECTION OPERATOR:
Least absolute shrinkage and selection operator (LASSO) is a regression analysis method that performs both variable selection and regularization to enhance the prediction accuracy and interpretability of the statistical model it produces. In our study, LASSO was utilized to evaluate the weight of each m6A regulator in the prognosis of patients [18]. The risk score of each patient was calculated with the following equation: risk score ∑i=1nCoef(i)*χ(i), where coef(i) was the value of each gene from LASSO analysis and x(i) was the expression value of each gene.
CONSENSUS CLUSTERING ANALYSIS:
The ConsensusClusterPlus package in R was used to perform consensus cluster analysis in our study. The algorithm started with subsetting a part of the characteristics and items in the data file, and each subset was divided into k groups according to k-means, the clustering algorithm used. This procedure was performed for a customized number of repetitions to yield a way to represent the consensus among several runs of the clustering and evaluate the solidity of the discovered clustering. Pairwise consensus values, the ratio of clustering runs in which 2 items are grouped together, were computed and kept in a consensus matrix for each k [19]. The clusters of patients with breast cancer were obtained according to the expression of m6A regulators. The prognosis of the different clusters was then compared.
ETHICS STATEMENT:
The study was approved by the institutional review board of our institution.
STATISTICS:
Chi-square tests or one-way ANOVA were applied to compare clinical and pathological data. Univariate and multivariate Cox regression analyses were conducted to evaluate the prognostic value of multiple parameters and characteristics. The Kaplan-Meier method was used to compare the OS of patients in different groups. All statistical analyses were conducted using R v3.6.1 (
Results
RNA EXPRESSION AND CLINICOPATHOLOGICAL CHARACTERISTICS:
The m6A regulators used in our study were collected from published studies, and 20 molecules were included. The Nottingham prognostic index (NPI) was used to predict the outcomes of breast cancers following surgery [20]. First, we evaluated the potential relationships between the RNA expression of m6A regulators and NPI, cancer cellularity, and tumor staging. As shown in Figure 1A, about half of the 20 m6A regulators were statistically significantly related with tumor staging. In particular, EIF3 expression increased as tumor staging increased from stage I to stage III (Figure 1B). By contrast, the gradually decreased expression of RBM15B was observed as tumor staging increased (Figure 1C). Also, the link of NPI to m6A regulators was determined. Interestingly, most of the genes were associated with the NPI value (Figure 1D). EIF3 and IGF2BP3 expression were further invalidated in different NPI groups. Results showed a growing trend of EIF3 expression as NPI value became greater (Figure 1E), and IGF2BP3 showed the same expression pattern as EIF3 (Figure 1F). In addition, the role of m6A regulators in cellularity was assessed. Those results indicated that more than half of these genes were involved in cellularity distribution at a statistically significant level (Figure 1G). EIF3 (Figure 1H) and RBM15 (Figure 1I) showed similar expression patterns, in which both decreased as the cellularity proportion shifted from high to low. We also displayed the expression of m6A regulators across different tumor stages, cellularity, and NPI scores in Supplementary Table 1.
:
We also performed consensus clustering to yield an appropriate k value that could perfectly separate patients with breast cancer. k=4 was selected as an adequate choice, with the stability of clustering rising from k = 2 to 10, according to the expression likeness of the m6A regulators (Figure 2A, 2B). From the k value of 4, patients were divided into 4 clusters based on the consensus clustering results. Survival analysis was then conducted to compare whether the 4 groups of patients showed distinct 10-year OS. As shown in Figure 2C, patients in different clusters had significantly different outcomes, indicating that the clustering was powerful enough to predict the prognosis of patients with breast cancer.
:
Because the regulators were all involved in m6A modification, they were expected to be closely connected with each other. We first searched the STRING database to evaluate the genes’ protein interaction with each other. As shown in Figure 3A, the identified genes in STRING were closely correlated, indicating that these m6A regulators were functionally interrelated at the protein level. Then, the correlation between the regulators’ RNA in the TCGA database was further evaluated. The results indicated that most of the RNAs were related with each other, although with a small coefficient (Figure 3B), which was not consistent with protein interactions or with the results from a previously published study [11]. This might be due to the tumor heterogeneity across different types of cancers or tumor purity. Because the TCGA dataset was not adjusted by tumor purity, we analyzed data from the TIMER database, in which gene expression is adjusted by tumor purity, to thoroughly measure the link among the regulators. With the TIMER database, we found some stronger associations among these regulators than with raw data from the TCGA database. Four pairs of regulators that were closely related to each other are shown in Figure 3C.
:
Based on the above findings, we believed that these regulators contributed to patients’ outcomes. Cox regression analysis was then used to estimate the role of these genes in the OS of breast cancer patients. The results showed that most of the modification regulators were related with poor OS and 5 of them could predict OS (P<0.05) (Figure 4A). LASSO analysis was then used to additionally assess the effect of the 5 genes on prognosis, and the risk coefficient was acquired for each of the 5 regulators (Figure 4B). The results were consistent with the Cox regression analysis, showing that the LASSO risk coefficient was not zero. Subsequently, patients were divided into 2 groups according to the median risk score, which was calculated with the following equation: risk score ∑i=1nCoef(i)*χ(i). The survival analysis indicated that the risk score could effectively separate patients’ survival (P<0.0001) (Figure 4C). Multivariate analysis also demonstrated that the risk score was an independent prognosis prediction factor (hazard ratio 1.23, 95% confidence interval [CI] 1.08–1.39; P=0.002) (Table 1). These results showed the strong power of m6A regulators in predicting prognosis of breast cancer. In addition, other independent prognosis prediction factors in our study were patient age, radiotherapy, HER2 status, positive lymph node number, and tumor size (Table 1). GSEA analysis was then conducted to explore the potential underlying mechanisms, pathways, and cancer hallmarks. As shown in Figure 4D, 4 common cancer-related pathways, AKT1 signaling, cytokinesis, RNA transcription, and APC-MYC pathway, were enriched in patients with a high-risk score. Next, we explored whether the risk score influenced the effect of radiotherapy, chemotherapy, and hormone therapy on breast cancer. First, we identified patients who had received chemotherapy, radiotherapy, or hormone therapy. Then, survival analysis was performed in these patients. The results suggested that patients with a high risk score had relatively worse survival compared with those with a low risk score (Figure 4E), indicating that high-risk cases were not as sensitive to chemoradiotherapy or hormone therapies as were low-risk cases. Therefore, we recommend that patients with a high risk should receive closer follow-up to aid in early diagnosis and treatment of recurrence.
Discussion
Prior work has documented the role of m6A regulators in breast carcinogenesis, development, and drug resistance; for example, researchers found that FTO can promote the progression of breast tumors [21] and HNRNPA2/B1 is altered in endocrine-resistant breast cancer [22]. However, these studies have either included a single m6A regulator or were not focused on clinical relevance. In the present study, we studied the m6A regulators as a whole in breast cancer to test their relevance with clinicopathological features.
We identified that the RNA expression of m6A regulators is closely related to prognosis and other clinical features such as the NPI and cellularity of breast cancer. Here, we first divided breast cancer patients into 4 clusters by consensus clustering and found that the clusters significantly affected the prognosis of patients with breast cancer. Also, patients were further split into 2 subgroups according to the risk score value. Risk was correlated with prognosis and influenced cancer crucial pathways and biological processes of breast cancer.
Twenty m6A regulators have been included in this study. These regulators were searched from previously published data and have been validated to be involved in m6A modification in various cancer types besides breast cancer. According to the theory of tumor heterogeneity [23], which states that different tumor cells and tissues of distinct morphological and phenotypic profiles exist in all cancers, the function of some m6A regulators in breast cancer might be different from that in other cancers, where the role of the m6A regulators have been validated. This could lead to some inconsistencies between our results and those of other studies. For example, WTAP was reported to promote genes in various cancers, including hepatocellular cancer [24], bladder cancer [25], and ovarian cancer [26]. However, in the present study, WTAP was found to be a protective factor of OS. The inconsistency could be the result of the limited sample size and gap between clinical and experimental differences in the present study. CBLL1 is mainly studied in lung cancer and has been recognized to promote cancer progression in lung cancer [27]. It was also related to a favorable prognosis in our study, which warrants more studies to explore the potential effect of CBLL1 in breast cancer and the tumor heterogeneity between lung cancer and breast cancer. Our findings also indicated that dysregulation of certain m6A regulators resulted in alterations of RNAs in cancers and validated that the same regulators might exert divergent functions in different tumors.
In our study, the relationship between m6A regulators and clinicopathological characteristics was also analyzed. As a “reader”, EIF3 could be directly recruited by m6A methylation, which is followed by a cap-independent translation in the cellular stress response [28]. EIF3 is closely linked to the tumor stage, NPI, and cellularity of breast cancer. Also, EIF3 was associated with poor survival of breast cancer in the present study, which is consistent with previous research showing that EIF3 contributes to poor survival of patients with HER2 (+) breast cancer [29] and triple-negative breast cancer [30]. These pieces of evidence indicate that EIF3 has a pivotal role of in breast cancer. In addition, most of the regulators were found to be associated with breast cancer staging, NPI, and cellularity in our study, which is consistent with previous studies that found m6A is widely involved in cancer progression and prognosis [31].
It has been demonstrated that FTO can regulate cell migration and invasion in breast cancer via the miR-181b-3p/ARL5B signaling pathway, and high FTO expression is associated with advanced breast cancer staging [32]. A recent study also showed that tumor status and stage were relevant to the expression level of m6A RNA methylation regulators [33]. The m6A in peripheral blood RNA is closely associated with breast cancer stage [34]. The regulators promote the invasiveness of breast cancer, and therefore are correlated with an advanced tumor stage. NPI is calculated based on 3 pathological criteria: tumor size, the number of involved lymph nodes, and tumor grade [20]. The m6A regulators are involved in lymph node metastasis [32] and are therefore associated with NPI. However, low expression of some m6A regulators, such as METTL14 and ZC3H13, is associated with tumor progression, including NPI [35]. This could be explained by the potential dual function of m6A regulators in breast cancer. We also searched data on the relationship between m6A and cellularity in breast cancer but found little evidence about that relationship. This indicates that our findings are novel and might provide some clues for future studies.
Chemotherapy, radiation, and hormone therapy have been shown to greatly improve the prognosis of breast cancer. In addition to predicting OS, the risk score also performs well when predicting the response of patients with breast cancer after receiving the above treatment modalities. Therefore, patients with a high risk score should receive a higher dose of adjuvant therapies and close surveillance after treatment. Preclinical studies have indicated that the activation of PI3K/AKT/mTOR contributes to an acquired resistance to hormone therapy [36,37]. Data from other randomized trials also demonstrated that mTOR inhibition could improve hormone therapy resistance [38,39]. It has been reported that MYC is frequently increased in chemotherapy-resistant breast cancer patients and that MYC is able to maintain breast cancer stem cells in chemotherapy-resistant patients [40]. High-risk breast cancer patients are harboring the enrichment of the above pathways, which probably leads to radiation, chemotherapy, and hormone therapy resistance. These molecules and hallmarks have been targeted to develop new drugs and overcome resistance to adjuvant therapies. Further studies targeting m6A methylation regulators are warranted.
Solid tumor tissues consist of cancerous and noncancerous components, which contain epithelial, stromal, and endothelial cells [41]. Tumor purity, characterized as the proportion of cancerous cells in a solid tumor sample, is a key feature in cancer transcriptomics and metabolomics data analysis. Noncancerous components are viewed as prevailing pollutants in commixture and constitute a massive part of tumor masses. Studies have found that m6A modulators are closely related to each other, and our results from the STRING and TIMER databases also invalidated the findings. However, the correlation result of RNA data from the TCGA database without tumor purity adjusted was not consistent with the results of STRING or TIMER, an inconsistency that could have resulted from tumor purity. Typically, harvested surgical samples could be of a purity of lower than 70%, giving rise to systematic biases in genomic analysis [42]. Tumor purity is an important confounder and therefore should be considered to increase data accuracy and reliability.
Conclusions
In conclusion, our data demonstrated that m6A regulators were related to clinicopathological characteristics and showed prognostic significance in breast cancer. Future studies to evaluate the role of m6A regulators in other types of cancer are needed. The development of targeted therapeutics based on m6A regulators is also promising.
Figures
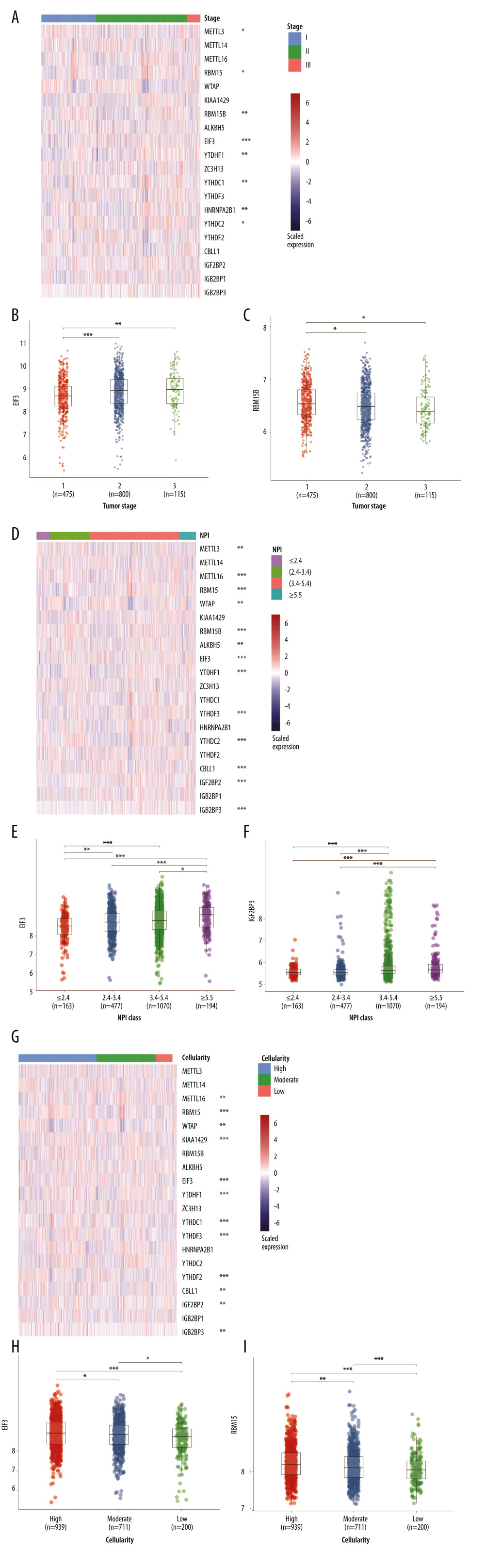 Figure 1. RNA expression and clinicopathological characteristics. (A) Heatmap of m6A regulators expression in patients with different tumor stages and significance. (B) EIF3 RNA expression in patients with stages I, II, and III. (C) RBM15B RNA expression in patients with stages I, II, and III. (D) Heatmap of m6A regulators expression in patients of different Nottingham prognostic index (NPI). (E) EIF3 RNA expression in patients with different NPI. (F) IGF2BP3 RNA expression patients with different NPI. (G) Heatmap of m6A regulators expression in patients of different cellularity. (H) EIF3 RNA expression in patients of different cellularity. (I) RBM15 RNA expression patients of different cellularity. * P<0.5, ** P<0.01, *** P<0.001.
Figure 1. RNA expression and clinicopathological characteristics. (A) Heatmap of m6A regulators expression in patients with different tumor stages and significance. (B) EIF3 RNA expression in patients with stages I, II, and III. (C) RBM15B RNA expression in patients with stages I, II, and III. (D) Heatmap of m6A regulators expression in patients of different Nottingham prognostic index (NPI). (E) EIF3 RNA expression in patients with different NPI. (F) IGF2BP3 RNA expression patients with different NPI. (G) Heatmap of m6A regulators expression in patients of different cellularity. (H) EIF3 RNA expression in patients of different cellularity. (I) RBM15 RNA expression patients of different cellularity. * P<0.5, ** P<0.01, *** P<0.001. 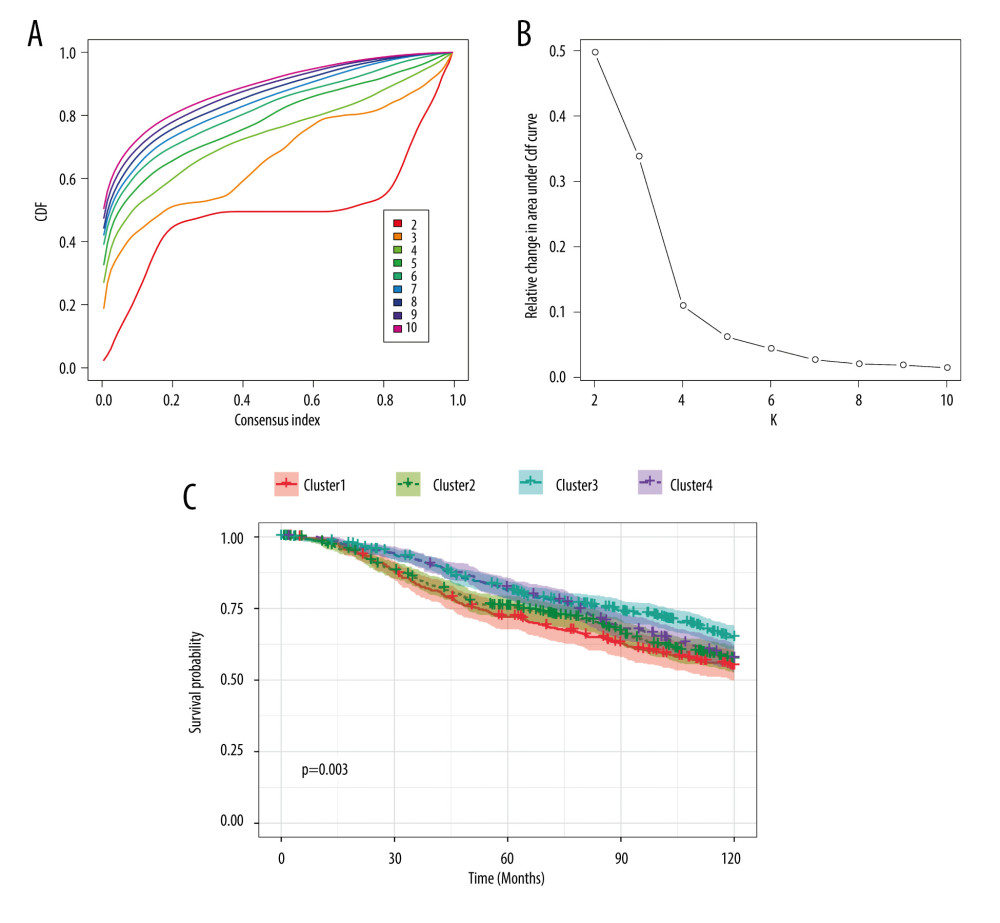 Figure 2. Consensus clustering of m6A regulators. (A) Consensus clustering cumulative distribution function (CDF) for k=2 to 10. (B) Relative change in area under CDF curve for k=2 to 10. (C) Overall survival analysis for breast cancer patients in 4 clusters.
Figure 2. Consensus clustering of m6A regulators. (A) Consensus clustering cumulative distribution function (CDF) for k=2 to 10. (B) Relative change in area under CDF curve for k=2 to 10. (C) Overall survival analysis for breast cancer patients in 4 clusters. 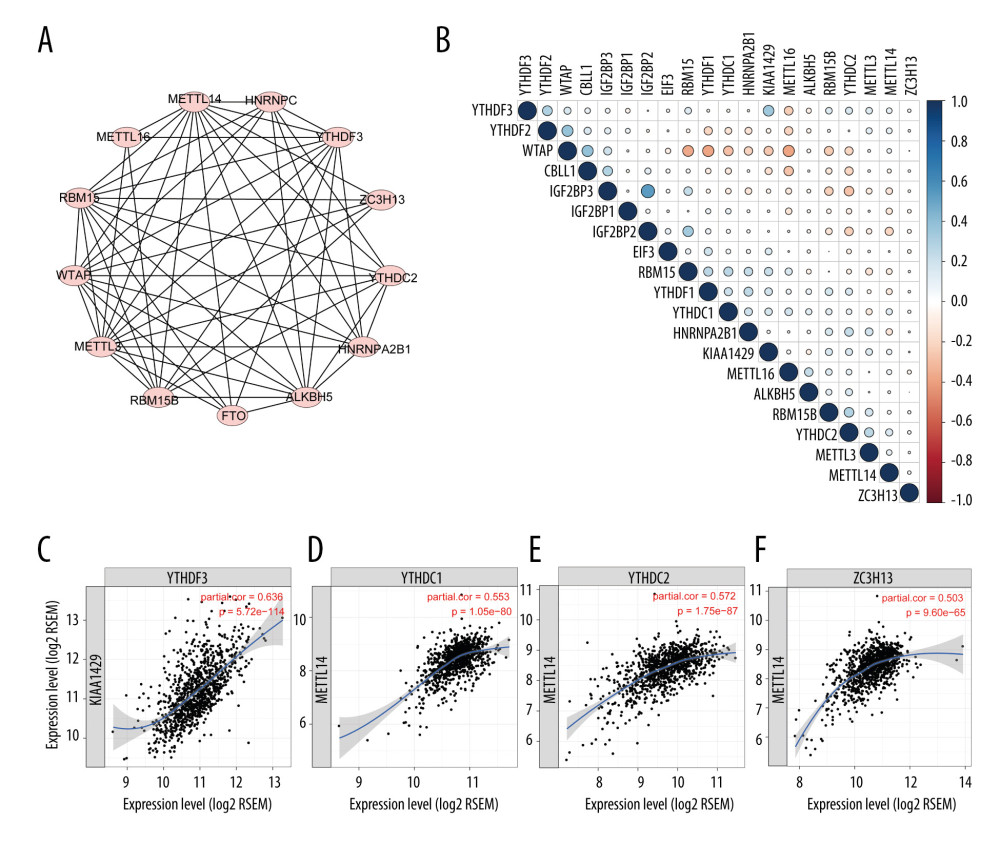 Figure 3. Interaction among m6A regulators. (A) Interactions across the m6A regulators from STRING database. (B) Correlation plot of m6A regulators RNA from the TCGA database. (C) Representative figures of correlations of m6A regulators adjusted by tumor purity from the TIMER database.
Figure 3. Interaction among m6A regulators. (A) Interactions across the m6A regulators from STRING database. (B) Correlation plot of m6A regulators RNA from the TCGA database. (C) Representative figures of correlations of m6A regulators adjusted by tumor purity from the TIMER database. 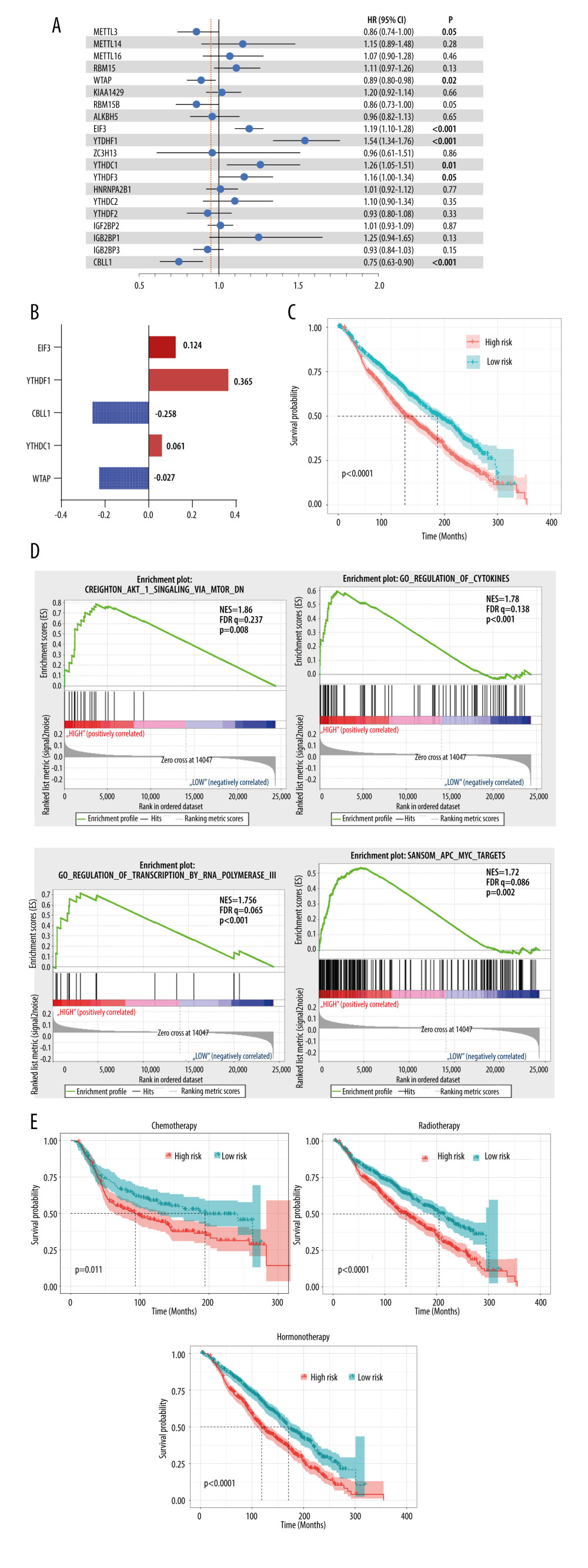 Figure 4. Clinical relevance of m6A regulators. (A) Forest plot of univariate Cox regression results of m6A regulators. (B) Coefficients calculated by multivariate Cox regression using Least absolute shrinkage and selection operator are shown. (C) Kaplan-Meier overall survival curves of patients that were divided into 2 groups based on the median risk score. (D) Gene set enrichment analysis results of patients in the high-risk group versus low-risk group. (E) Kaplan-Meier overall survival curves of patients that received chemoradiotherapy or hormone therapy.
Figure 4. Clinical relevance of m6A regulators. (A) Forest plot of univariate Cox regression results of m6A regulators. (B) Coefficients calculated by multivariate Cox regression using Least absolute shrinkage and selection operator are shown. (C) Kaplan-Meier overall survival curves of patients that were divided into 2 groups based on the median risk score. (D) Gene set enrichment analysis results of patients in the high-risk group versus low-risk group. (E) Kaplan-Meier overall survival curves of patients that received chemoradiotherapy or hormone therapy. References
1. Siegel RL, Miller KD, Jemal A, Cancer statistics, 2020: Cancer J Clin, 2020; 70(1); 7-30
2. Woolston C, Breast cancer: 4 big questions: Nature, 2015; 527(7578); S120
3. Zhao Y, Shi Y, Shen H, Xie W, m(6)A-binding proteins: the emerging crucial performers in epigenetics: J Hematol Oncol, 2020; 13(1); 35
4. Nandy D, Rajam SM, Dutta D, A three layered histone epigenetics in breast cancer metastasis: Cell Biosci, 2020; 10; 52
5. Desrosiers R, Friderici K, Rottman F, Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells: Proc Natl Acad Sci USA, 1974; 71(10); 3971-75
6. Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR, RNA methylation by the MIS complex regulates a cell fate decision in yeast: PLoS Genet, 2012; 8(6); e1002732
7. Wang X, Feng J, Xue Y, Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex: Nature, 2016; 534(7608); 575-78
8. Zheng G, Dahl JA, Niu Y, ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility: Mol Cell, 2013; 49(1); 18-29
9. Jia G, Fu Y, Zhao X, N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO: Nat Chem Biol, 2011; 7(12); 885-87
10. Yang Y, Hsu PJ, Chen YS, Yang YG, Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism: Cell Res, 2018; 28(6); 616-24
11. Li Y, Xiao J, Bai J, Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types: Mol Cancer, 2019; 18(1); 137
12. Lan Q, Liu PY, Haase J, The critical role of RNA m(6)A methylation in cancer: Cancer Res, 2019; 79(7); 1285-92
13. Wang H, Xu B, Shi J, N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2: Gene, 2020; 722; 144076
14. Liu L, Liu X, Dong Z, N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival: J Cancer, 2019; 10(22); 5447-59
15. Szklarczyk D, Gable AL, Lyon D, STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets: Nucleic Acids Res, 2019; 47(D1); D607-13
16. Hsiao KY, Lin YC, Gupta SK, Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis: Cancer Res, 2017; 77(9); 2339-50
17. Subramanian A, Kuehn H, Gould J, GSEA-P: A desktop application for Gene Set Enrichment Analysis: Bioinformatics, 2007; 23(23); 3251-53
18. Chai RC, Wu F, Wang QX, m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas: Aging (Albany NY), 2019; 11(4); 1204-25
19. Zhang S, Wang Y, Gu Y, Specific breast cancer prognosis-subtype distinctions based on DNA methylation patterns: Mol Oncol, 2018; 12(7); 1047-60
20. Haybittle JL, Blamey RW, Elston CW, A prognostic index in primary breast cancer: Br J Cancer, 1982; 45(3); 361-66
21. Niu Y, Lin Z, Wan A, RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3: Mol Cancer, 2019; 18(1); 46
22. Klinge CM, Piell KM, Tooley CS, Rouchka EC, HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells: Sci Rep, 2019; 9(1); 9430
23. Marusyk A, Polyak K, Tumor heterogeneity: causes and consequences: Biochim Biophys Acta, 2010; 1805(1); 105-17
24. Chen Y, Peng C, Chen J, WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1: Mol Cancer, 2019; 18(1); 127
25. Chen L, Wang X, Relationship between the genetic expression of WTAP and bladder cancer and patient prognosis: Oncol Lett, 2018; 16(6); 6966-70
26. Yu HL, Ma XD, Tong JF, WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells: Onco Targets Ther, 2019; 12; 6191-201
27. Hui L, Zhang S, Wudu M, CBLL1 is highly expressed in non-small cell lung cancer and promotes cell proliferation and invasion: Thorac Cancer, 2019; 10(6); 1479-88
28. Meyer KD, Patil DP, Zhou J, 5′ UTR m(6)A promotes cap-independent translation: Cell, 2015; 163(4); 999-1010
29. Kar A, Gutierrez-Hartmann A, ESE-1/ELF3 mRNA expression associates with poor survival outcomes in HER2(+) breast cancer patients and is critical for tumorigenesis in HER2(+) breast cancer cells: Oncotarget, 2017; 8(41); 69622-40
30. Zhao ZM, Yost SE, Hutchinson KE, CCNE1 amplification is associated with poor prognosis in patients with triple negative breast cancer: BMC Cancer, 2019; 19(1); 96
31. Chen XY, Zhang J, Zhu JS, The role of m(6)A RNA methylation in human cancer: Mol Cancer, 2019; 18(1); 103
32. Xu Y, Ye S, Zhang N, The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer: Cancer Commun (Lond), 2020; 40(10); 484-500
33. Zhang B, Gu Y, Jiang G, Expression and prognostic characteristics of m(6) A RNA methylation regulators in breast cancer: Front Genet, 2020; 11; 604597
34. Xiao H, Fan X, Zhang R, Wu G, Upregulated N6-methyladenosine RNA in peripheral blood: potential diagnostic biomarker for breast cancer: Cancer Res Treat, 2021; 53(2); 399-408
35. Gong PJ, Shao YC, Yang Y, Analysis of N6-methyladenosine methyltransferase reveals METTL14 and ZC3H13 as tumor suppressor genes in breast cancer: Front Oncol, 2020; 10; 578963
36. Miller TW, Hennessy BT, Gonzalez-Angulo AM, Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer: J Clin Invest, 2010; 120(7); 2406-13
37. Sanchez CG, Ma CX, Crowder RJ, Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer: Breast Cancer Res, 2011; 13(2); R21
38. Bachelot T, Bourgier C, Cropet C, Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer with prior exposure to aromatase inhibitors: a GINECO study: J Clin Oncol, 2012; 30(22); 2718-24
39. Baselga J, Campone M, Piccart M, Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer: N Engl J Med, 2012; 366(6); 520-29
40. Lee KM, Giltnane JM, Balko JM, MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation: Cell Metab, 2017; 26(4); 633-47.e7
41. Rhee JK, Jung YC, Kim KR, Impact of tumor purity on immune gene expression and clustering analyses across multiple cancer types: Cancer Immunol Res, 2018; 6(1); 87-97
42. Aran D, Sirota M, Butte AJ, Systematic pan-cancer analysis of tumour purity: Nat Commun, 2015; 6; 8971
Figures
 Figure 1. RNA expression and clinicopathological characteristics. (A) Heatmap of m6A regulators expression in patients with different tumor stages and significance. (B) EIF3 RNA expression in patients with stages I, II, and III. (C) RBM15B RNA expression in patients with stages I, II, and III. (D) Heatmap of m6A regulators expression in patients of different Nottingham prognostic index (NPI). (E) EIF3 RNA expression in patients with different NPI. (F) IGF2BP3 RNA expression patients with different NPI. (G) Heatmap of m6A regulators expression in patients of different cellularity. (H) EIF3 RNA expression in patients of different cellularity. (I) RBM15 RNA expression patients of different cellularity. * P<0.5, ** P<0.01, *** P<0.001.
Figure 1. RNA expression and clinicopathological characteristics. (A) Heatmap of m6A regulators expression in patients with different tumor stages and significance. (B) EIF3 RNA expression in patients with stages I, II, and III. (C) RBM15B RNA expression in patients with stages I, II, and III. (D) Heatmap of m6A regulators expression in patients of different Nottingham prognostic index (NPI). (E) EIF3 RNA expression in patients with different NPI. (F) IGF2BP3 RNA expression patients with different NPI. (G) Heatmap of m6A regulators expression in patients of different cellularity. (H) EIF3 RNA expression in patients of different cellularity. (I) RBM15 RNA expression patients of different cellularity. * P<0.5, ** P<0.01, *** P<0.001. Figure 2. Consensus clustering of m6A regulators. (A) Consensus clustering cumulative distribution function (CDF) for k=2 to 10. (B) Relative change in area under CDF curve for k=2 to 10. (C) Overall survival analysis for breast cancer patients in 4 clusters.
Figure 2. Consensus clustering of m6A regulators. (A) Consensus clustering cumulative distribution function (CDF) for k=2 to 10. (B) Relative change in area under CDF curve for k=2 to 10. (C) Overall survival analysis for breast cancer patients in 4 clusters. Figure 3. Interaction among m6A regulators. (A) Interactions across the m6A regulators from STRING database. (B) Correlation plot of m6A regulators RNA from the TCGA database. (C) Representative figures of correlations of m6A regulators adjusted by tumor purity from the TIMER database.
Figure 3. Interaction among m6A regulators. (A) Interactions across the m6A regulators from STRING database. (B) Correlation plot of m6A regulators RNA from the TCGA database. (C) Representative figures of correlations of m6A regulators adjusted by tumor purity from the TIMER database. Figure 4. Clinical relevance of m6A regulators. (A) Forest plot of univariate Cox regression results of m6A regulators. (B) Coefficients calculated by multivariate Cox regression using Least absolute shrinkage and selection operator are shown. (C) Kaplan-Meier overall survival curves of patients that were divided into 2 groups based on the median risk score. (D) Gene set enrichment analysis results of patients in the high-risk group versus low-risk group. (E) Kaplan-Meier overall survival curves of patients that received chemoradiotherapy or hormone therapy.
Figure 4. Clinical relevance of m6A regulators. (A) Forest plot of univariate Cox regression results of m6A regulators. (B) Coefficients calculated by multivariate Cox regression using Least absolute shrinkage and selection operator are shown. (C) Kaplan-Meier overall survival curves of patients that were divided into 2 groups based on the median risk score. (D) Gene set enrichment analysis results of patients in the high-risk group versus low-risk group. (E) Kaplan-Meier overall survival curves of patients that received chemoradiotherapy or hormone therapy. Tables
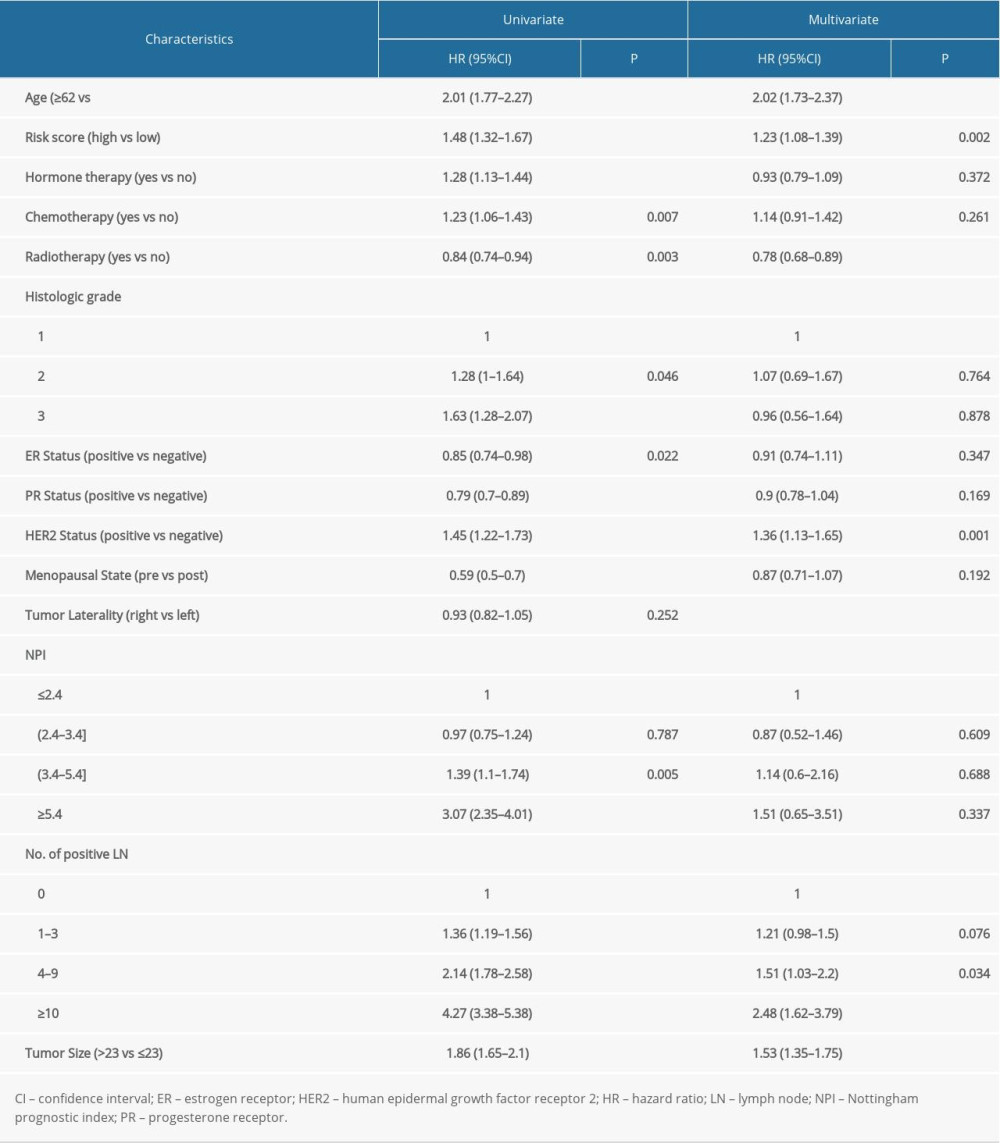 Table 1. Cox regression analysis of risk scores and clinicopathological factors for patients’ overall survival.
Table 1. Cox regression analysis of risk scores and clinicopathological factors for patients’ overall survival. Table 1. Cox regression analysis of risk scores and clinicopathological factors for patients’ overall survival.
Table 1. Cox regression analysis of risk scores and clinicopathological factors for patients’ overall survival.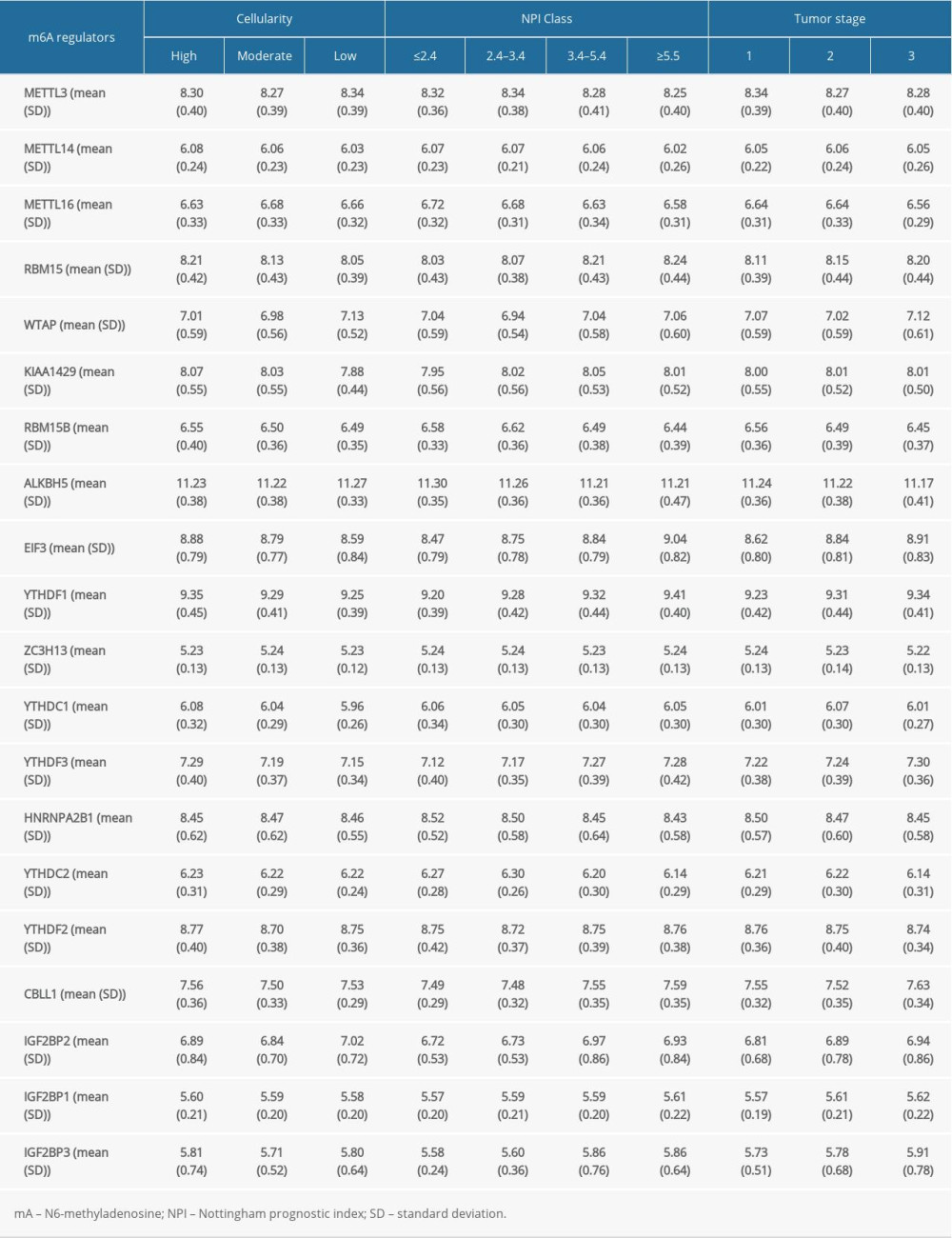 Supplementary Table 1. Expression of N6-methyladenosine regulators in different cellularity, tumor stage, or Nottingham prognostic index groups.
Supplementary Table 1. Expression of N6-methyladenosine regulators in different cellularity, tumor stage, or Nottingham prognostic index groups. In Press
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








